NPs Basic Information
|
Name |
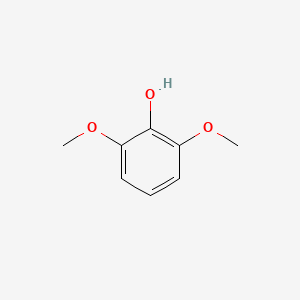
2,6-Dimethoxyphenol
|
| Molecular Formula | C8H10O3 | |
| IUPAC Name* |
2,6-dimethoxyphenol
|
|
| SMILES |
COC1=C(C(=CC=C1)OC)O
|
|
| InChI |
InChI=1S/C8H10O3/c1-10-6-4-3-5-7(11-2)8(6)9/h3-5,9H,1-2H3
|
|
| InChIKey |
KLIDCXVFHGNTTM-UHFFFAOYSA-N
|
|
| Synonyms |
2,6-DIMETHOXYPHENOL; 91-10-1; Syringol; Pyrogallol 1,3-dimethyl ether; Phenol, 2,6-dimethoxy-; 1,3-Dimethoxy-2-hydroxybenzene; 2-Hydroxy-1,3-dimethoxybenzene; 1,3-Dimethyl pyrogallate; 2,6-dimethoxy phenol; Aldrich; 1,3-Di-o-methylpyrogallol; 2,6-dimethoxy-phenol; Pyrogallol dimethylether; 2,6-Dwumetoksyfenol; FEMA No. 3137; 2,6-Dimethoxyphenol 99+%; 2,6-Dimethoxyphenyl; CHEBI:955; 4UQT464H8K; 33-51-2; 2,6-Dimethoxyphenol(Chunk or Granule or Flakes); 25511-61-9; 2,6-Dwumetoksyfenol [Polish]; EINECS 202-041-1; MFCD00064434; UNII-4UQT464H8K; 3DM; 2,6-dimetoxyphenol; 2,6,Dimethoxyphenol; 2,6-di-methoxyphenol; 1,3-dimethoxypyrogallol; bmse010203; DSSTox_CID_31180; DSSTox_GSID_52607; 2,6-Dimethoxyphenol, 99%; SCHEMBL156388; CHEMBL109652; 2,6-Dimethoxyphenol (syringol); DTXSID2052607; FEMA 3137; ZINC154666; 2,6-DIMETHOXYPHENOL [FHFI]; Tox21_303953; 2,6-DIMETHOXY PHENOL [FCC]; BDBM50409535; 2,6-Dimethoxyphenol, >=98%, FG; AKOS000120263; CS-W003972; FS-1188; HY-W003972; 2,6-Dimethoxyphenol, natural, >=96%; CAS-91-10-1; 2,6-Dimethoxyphenol, analytical standard; NCGC00357187-01; BP-10363; DB-003242; D0639; FT-0631436; EN300-20299; F13351; 064D434; A843720; Q421420; 2,6-Dimethoxyphenol Pyrogallol 1,3-dimethyl ether; Q-200214; Z104477654
|
|
| CAS | 91-10-1 | |
| PubChem CID | 7041 | |
| ChEMBL ID | CHEMBL109652 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 154.16 | ALogp: | 1.1 |
| HBD: | 1 | HBA: | 3 |
| Rotatable Bonds: | 2 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 38.7 | Aromatic Rings: | 1 |
| Heavy Atoms: | 11 | QED Weighted: | 0.707 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.532 | MDCK Permeability: | 0.00002490 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.023 |
| Human Intestinal Absorption (HIA): | 0.004 | 20% Bioavailability (F20%): | 0.004 |
| 30% Bioavailability (F30%): | 0.013 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.664 | Plasma Protein Binding (PPB): | 79.53% |
| Volume Distribution (VD): | 0.978 | Fu: | 22.53% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.709 | CYP1A2-substrate: | 0.943 |
| CYP2C19-inhibitor: | 0.404 | CYP2C19-substrate: | 0.884 |
| CYP2C9-inhibitor: | 0.066 | CYP2C9-substrate: | 0.814 |
| CYP2D6-inhibitor: | 0.063 | CYP2D6-substrate: | 0.908 |
| CYP3A4-inhibitor: | 0.08 | CYP3A4-substrate: | 0.369 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 11.517 | Half-life (T1/2): | 0.908 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.051 | Human Hepatotoxicity (H-HT): | 0.085 |
| Drug-inuced Liver Injury (DILI): | 0.052 | AMES Toxicity: | 0.031 |
| Rat Oral Acute Toxicity: | 0.164 | Maximum Recommended Daily Dose: | 0.033 |
| Skin Sensitization: | 0.876 | Carcinogencity: | 0.697 |
| Eye Corrosion: | 0.94 | Eye Irritation: | 0.985 |
| Respiratory Toxicity: | 0.359 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000501 | 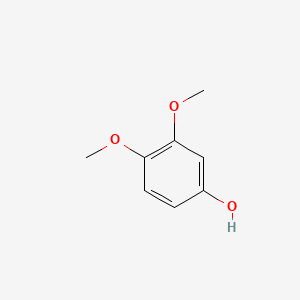 |
0.579 | D09GYT | 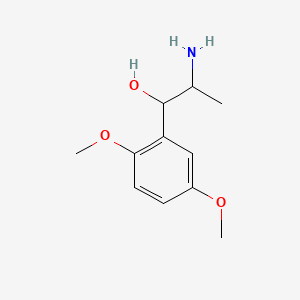 |
0.373 | ||
| ENC001512 | 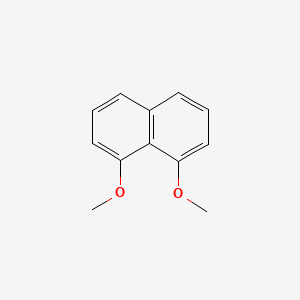 |
0.522 | D0E9CD |  |
0.364 | ||
| ENC004820 |  |
0.481 | D06TQZ |  |
0.333 | ||
| ENC000304 |  |
0.477 | D0FN7J | 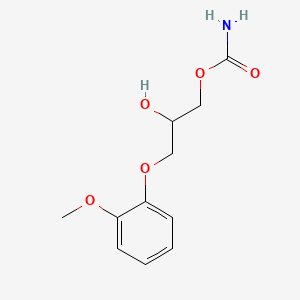 |
0.328 | ||
| ENC004830 | 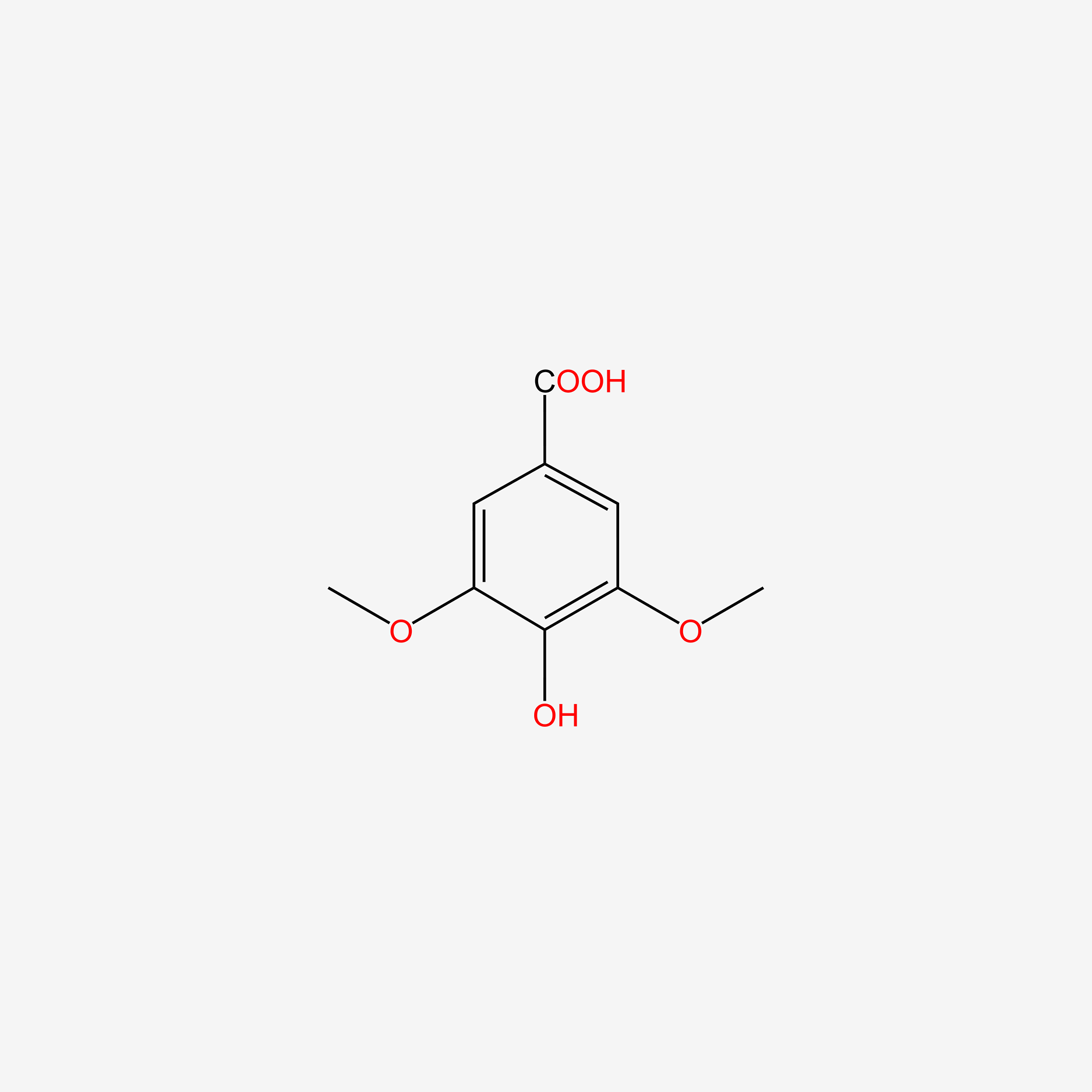 |
0.457 | D02XJY | 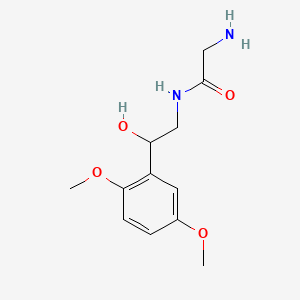 |
0.317 | ||
| ENC000367 | 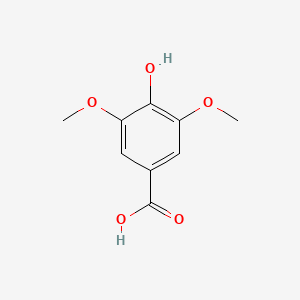 |
0.457 | D0A3HB | 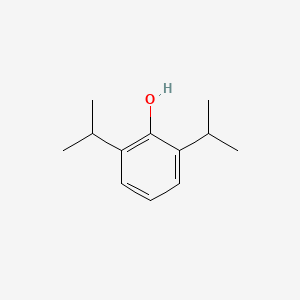 |
0.306 | ||
| ENC000033 | 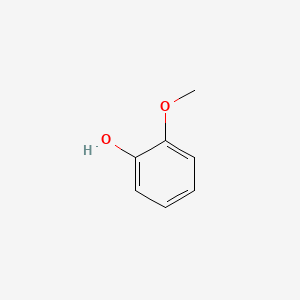 |
0.447 | D06GCK |  |
0.303 | ||
| ENC002213 | 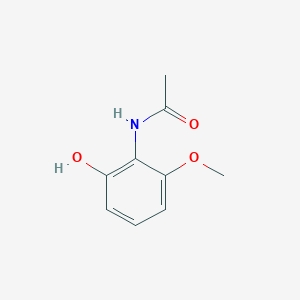 |
0.444 | D0Q9ON | 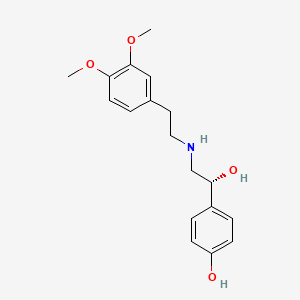 |
0.301 | ||
| ENC000478 | 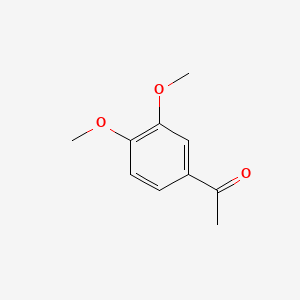 |
0.444 | D01SAT | 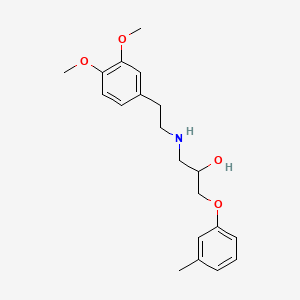 |
0.295 | ||
| ENC000712 | 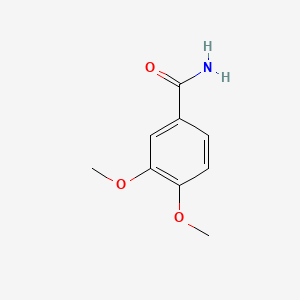 |
0.444 | D0E6OC | 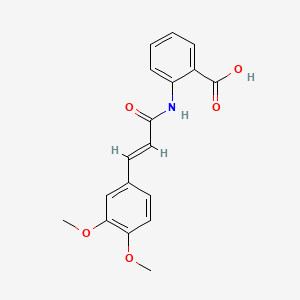 |
0.293 | ||