NPs Basic Information
|
Name |
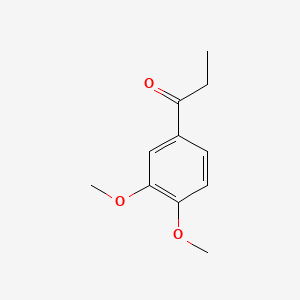
1-(3,4-Dimethoxyphenyl)propan-1-one
|
| Molecular Formula | C11H14O3 | |
| IUPAC Name* |
1-(3,4-dimethoxyphenyl)propan-1-one
|
|
| SMILES |
CCC(=O)C1=CC(=C(C=C1)OC)OC
|
|
| InChI |
InChI=1S/C11H14O3/c1-4-9(12)8-5-6-10(13-2)11(7-8)14-3/h5-7H,4H2,1-3H3
|
|
| InChIKey |
SBMSBQOMJGZBRY-UHFFFAOYSA-N
|
|
| Synonyms |
1-(3,4-dimethoxyphenyl)propan-1-one; 1835-04-7; 3,4-Dimethoxypropiophenone; Propioveratrone; 1-(3,4-Dimethoxy-phenyl)-propan-1-one; 1-Propanone, 1-(3,4-dimethoxyphenyl)-; 1-(3,4-Dimethoxyphenyl)-1-propanone; 3,4-Dimethoxyphenyl ethyl ketone; Propiophenone, 3',4'-dimethoxy-; 3',4'-DIMETHOXYPROPIOPHENONE; 3',4'-dimethoxy-1-phenylpropiophenone; MFCD00482089; AI3-23454; NSC16954; ghl.PD_Mitscher_leg0.580; MLS001048910; SCHEMBL515746; CHEMBL479685; DTXSID60171427; CHEBI:167438; HMS2268J04; ZINC370286; BBL009808; NSC 16954; NSC-16954; STK043726; AKOS000115379; 1-(3,4-dimethoxyphenyl)-propan-1-one; NCGC00246237-01; NS-01887; SMR000387103; Propan-1-one, 1-(3,4-dimethoxyphenyl)-; AM20030211; CS-0152658; FT-0614354; EN300-01082; Y14672; 1-(3,4-dimethoxy-phenyl)-propan-1-one, AldrichCPR; Z57952774
|
|
| CAS | 1835-04-7 | |
| PubChem CID | 15781 | |
| ChEMBL ID | CHEMBL479685 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 194.23 | ALogp: | 2.2 |
| HBD: | 0 | HBA: | 3 |
| Rotatable Bonds: | 4 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 35.5 | Aromatic Rings: | 1 |
| Heavy Atoms: | 14 | QED Weighted: | 0.691 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.524 | MDCK Permeability: | 0.00002570 |
| Pgp-inhibitor: | 0.024 | Pgp-substrate: | 0.033 |
| Human Intestinal Absorption (HIA): | 0.003 | 20% Bioavailability (F20%): | 0.004 |
| 30% Bioavailability (F30%): | 0.027 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.959 | Plasma Protein Binding (PPB): | 72.94% |
| Volume Distribution (VD): | 0.648 | Fu: | 16.67% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.935 | CYP1A2-substrate: | 0.95 |
| CYP2C19-inhibitor: | 0.583 | CYP2C19-substrate: | 0.846 |
| CYP2C9-inhibitor: | 0.112 | CYP2C9-substrate: | 0.714 |
| CYP2D6-inhibitor: | 0.048 | CYP2D6-substrate: | 0.872 |
| CYP3A4-inhibitor: | 0.109 | CYP3A4-substrate: | 0.459 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 9.316 | Half-life (T1/2): | 0.855 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.059 | Human Hepatotoxicity (H-HT): | 0.047 |
| Drug-inuced Liver Injury (DILI): | 0.335 | AMES Toxicity: | 0.032 |
| Rat Oral Acute Toxicity: | 0.099 | Maximum Recommended Daily Dose: | 0.025 |
| Skin Sensitization: | 0.155 | Carcinogencity: | 0.193 |
| Eye Corrosion: | 0.036 | Eye Irritation: | 0.635 |
| Respiratory Toxicity: | 0.421 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000478 | 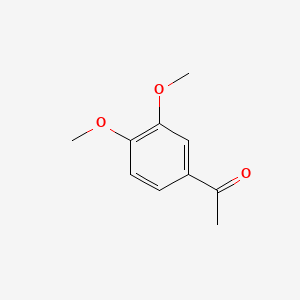 |
0.738 | D02XJY | 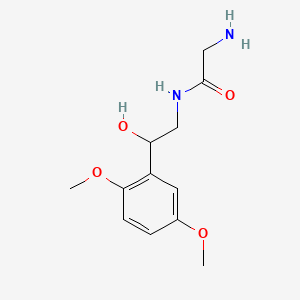 |
0.381 | ||
| ENC000712 | 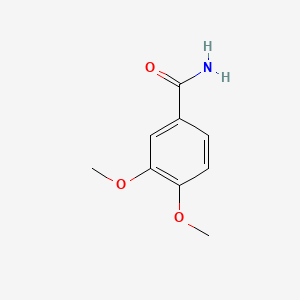 |
0.698 | D09GYT | 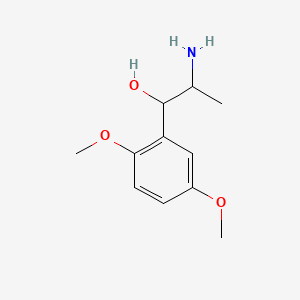 |
0.368 | ||
| ENC000501 | 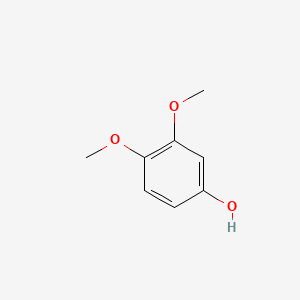 |
0.545 | D0VU8Q | 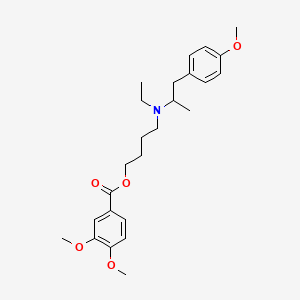 |
0.359 | ||
| ENC001363 | 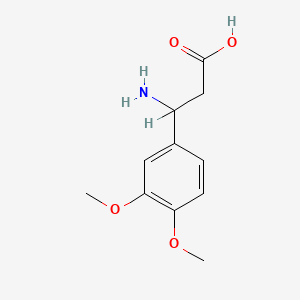 |
0.528 | D0E6OC | 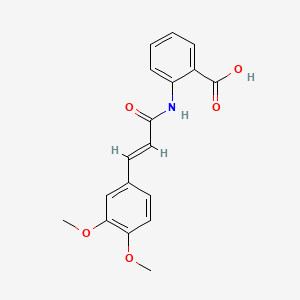 |
0.346 | ||
| ENC001461 | 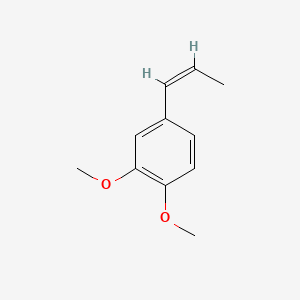 |
0.510 | D0E9CD | 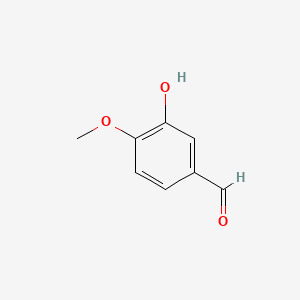 |
0.333 | ||
| ENC000777 | 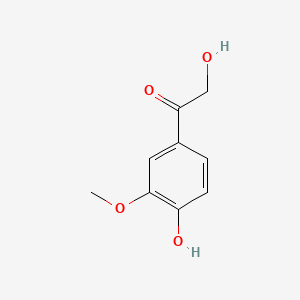 |
0.490 | D0L0ZF | 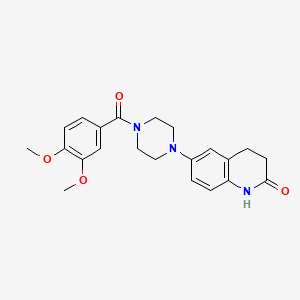 |
0.330 | ||
| ENC000367 | 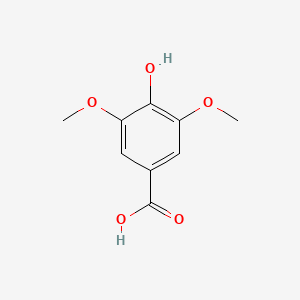 |
0.442 | D0F4ZY |  |
0.322 | ||
| ENC004830 | 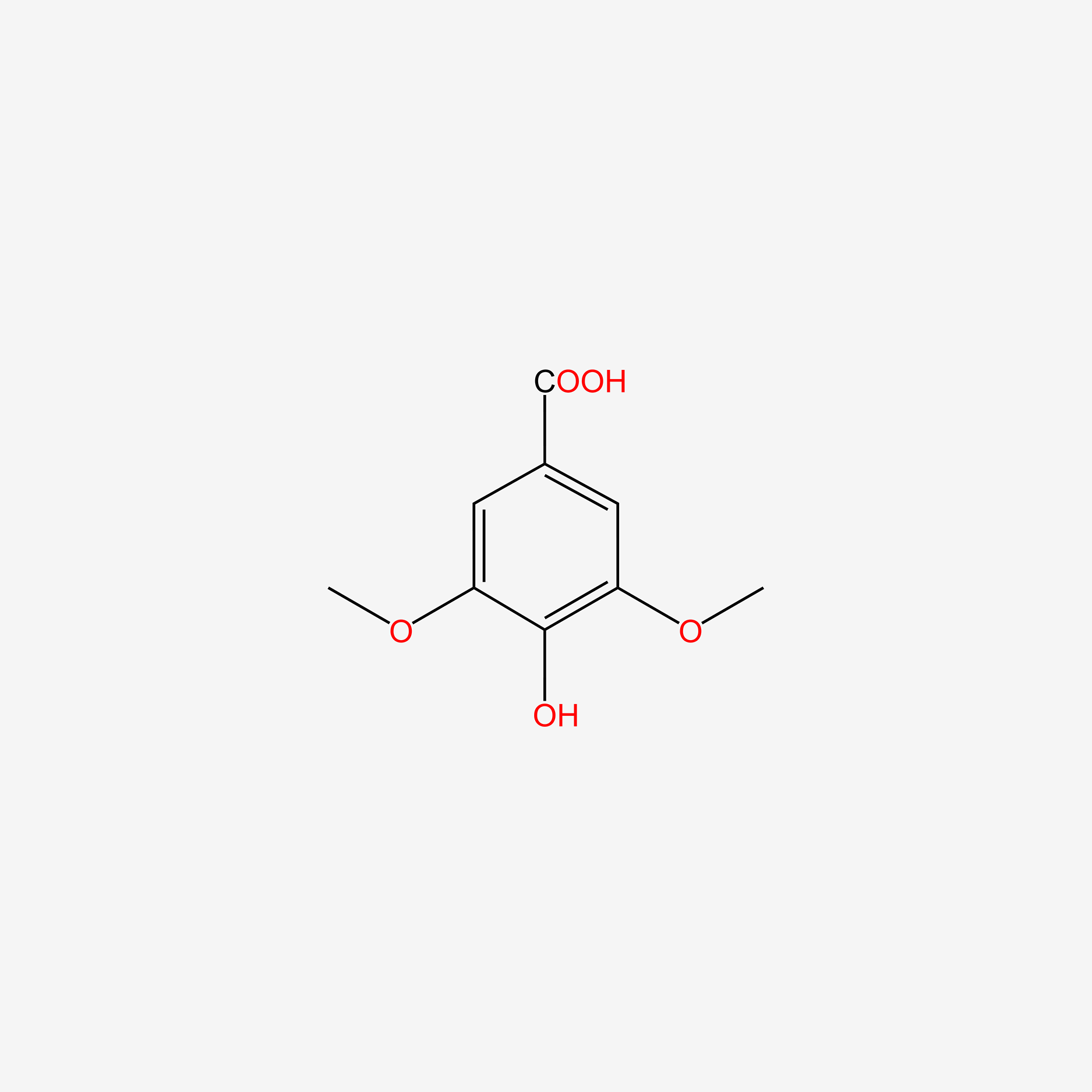 |
0.442 | D06GCK |  |
0.321 | ||
| ENC000296 | 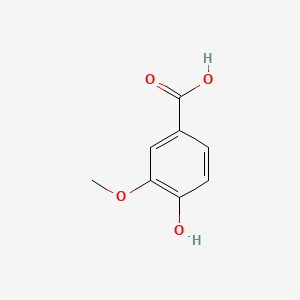 |
0.429 | D0Q9ON | 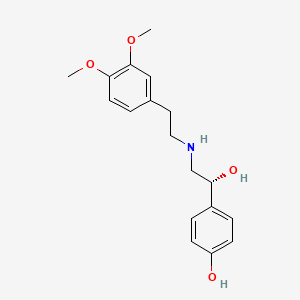 |
0.321 | ||
| ENC001056 | 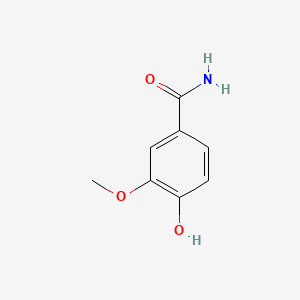 |
0.429 | D0A8FB | 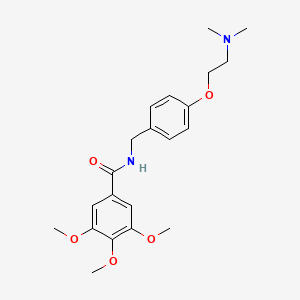 |
0.318 | ||