NPs Basic Information
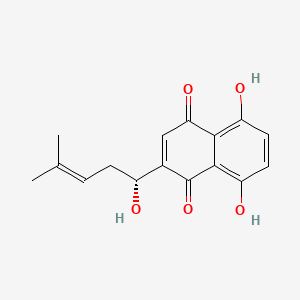
|
Name |
(+)-Shikonin
|
| Molecular Formula | C16H16O5 | |
| IUPAC Name* |
5,8-dihydroxy-2-[(1R)-1-hydroxy-4-methylpent-3-enyl]naphthalene-1,4-dione
|
|
| SMILES |
CC(=CC[C@H](C1=CC(=O)C2=C(C=CC(=C2C1=O)O)O)O)C
|
|
| InChI |
InChI=1S/C16H16O5/c1-8(2)3-4-10(17)9-7-13(20)14-11(18)5-6-12(19)15(14)16(9)21/h3,5-7,10,17-19H,4H2,1-2H3/t10-/m1/s1
|
|
| InChIKey |
NEZONWMXZKDMKF-SNVBAGLBSA-N
|
|
| Synonyms |
shikonin; 517-89-5; (R)-5,8-Dihydroxy-2-(1-hydroxy-4-methylpent-3-en-1-yl)naphthalene-1,4-dione; Isoarnebin 4; Tokyo Violet; (+)-Shikonin; 5,8-Dihydroxy-2-[(1R)-1-hydroxy-4-methyl-pent-3-enyl]naphthalene-1,4-dione; NSC 252844; Shikonine; CHEBI:81068; 5,8-dihydroxy-2-[(1R)-1-hydroxy-4-methylpent-3-enyl]naphthalene-1,4-dione; C.I. 75535; Shikonin S; 5,8-dihydroxy-2-[(1R)-1-hydroxy-4-methylpent-3-en-1-yl]-1,4-dihydronaphthalene-1,4-dione; SR-05000001466; UNII-3IK6592UBW; CCRIS 6492; HSDB 8100; (R)-Shikonin; Shikonin,(S); BRN 2058010; SCHEMBL33969; BSPBio_001270; MLS006010149; DTXSID30199653; HMS1792P11; HMS1990P11; HMS2089L09; HMS3403P11; AMY39404; HY-N0822; ZINC2015152; MFCD00075680; s8279; (+)-5,8-Dihydroxy-2-(1-hydroxy-4-methyl-3-pentenyl)-1,4-naphthoquinone; AKOS015900416; CCG-208272; CS-5906; NCGC00163489-01; NCGC00163489-02; NCGC00163489-04; NCGC00163489-05; AC-26871; AS-13554; BP-30212; SMR002530673; C15906; 517S895; J-502235; SR-05000001466-1; SR-05000001466-3; Q27155024
|
|
| CAS | 517-89-5 | |
| PubChem CID | 479503 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 288.29 | ALogp: | 3.0 |
| HBD: | 3 | HBA: | 5 |
| Rotatable Bonds: | 3 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 94.8 | Aromatic Rings: | 2 |
| Heavy Atoms: | 21 | QED Weighted: | 0.587 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.909 | MDCK Permeability: | 0.00000832 |
| Pgp-inhibitor: | 0.01 | Pgp-substrate: | 0.004 |
| Human Intestinal Absorption (HIA): | 0.021 | 20% Bioavailability (F20%): | 0.847 |
| 30% Bioavailability (F30%): | 0.99 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.005 | Plasma Protein Binding (PPB): | 96.12% |
| Volume Distribution (VD): | 0.681 | Fu: | 5.41% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.953 | CYP1A2-substrate: | 0.173 |
| CYP2C19-inhibitor: | 0.259 | CYP2C19-substrate: | 0.055 |
| CYP2C9-inhibitor: | 0.719 | CYP2C9-substrate: | 0.741 |
| CYP2D6-inhibitor: | 0.716 | CYP2D6-substrate: | 0.235 |
| CYP3A4-inhibitor: | 0.149 | CYP3A4-substrate: | 0.096 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 17.16 | Half-life (T1/2): | 0.783 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.008 | Human Hepatotoxicity (H-HT): | 0.264 |
| Drug-inuced Liver Injury (DILI): | 0.805 | AMES Toxicity: | 0.766 |
| Rat Oral Acute Toxicity: | 0.442 | Maximum Recommended Daily Dose: | 0.294 |
| Skin Sensitization: | 0.922 | Carcinogencity: | 0.454 |
| Eye Corrosion: | 0.033 | Eye Irritation: | 0.942 |
| Respiratory Toxicity: | 0.401 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC002125 | 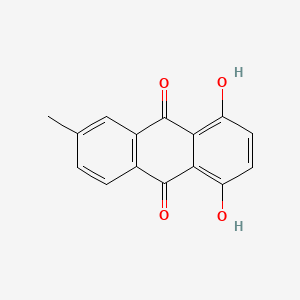 |
0.403 | D04PHC | 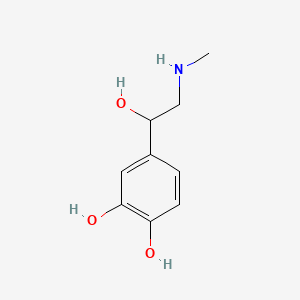 |
0.300 | ||
| ENC004048 | 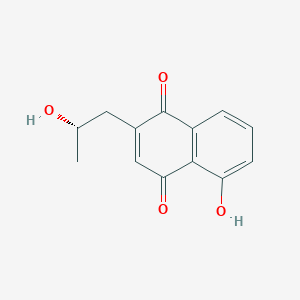 |
0.397 | D0I8FI | 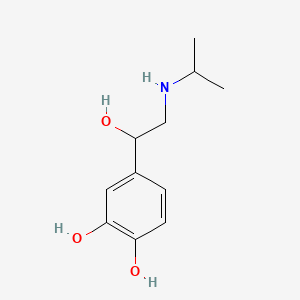 |
0.297 | ||
| ENC005353 | 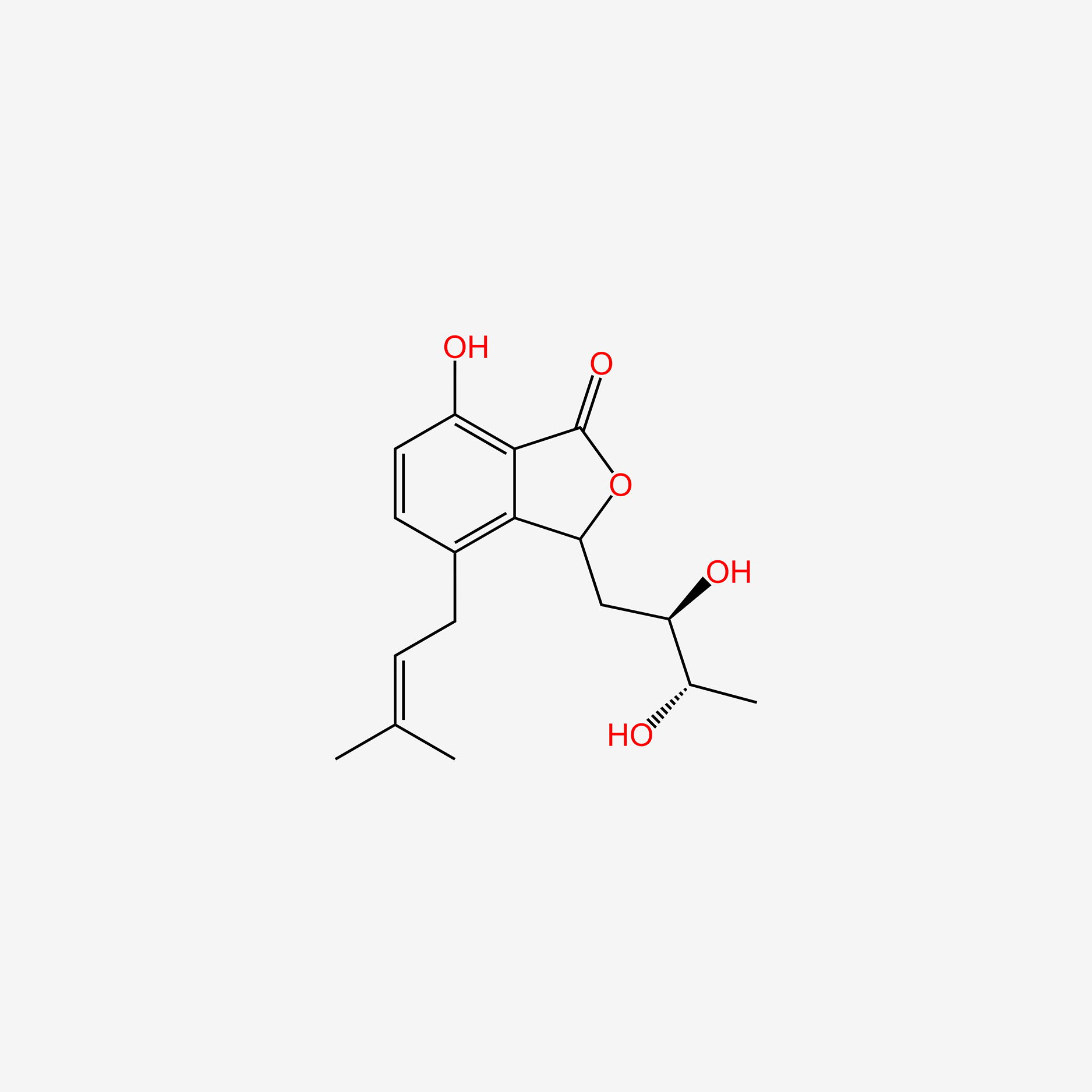 |
0.369 | D08FPM | 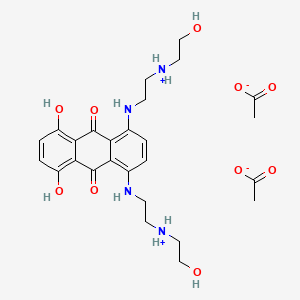 |
0.296 | ||
| ENC005576 | 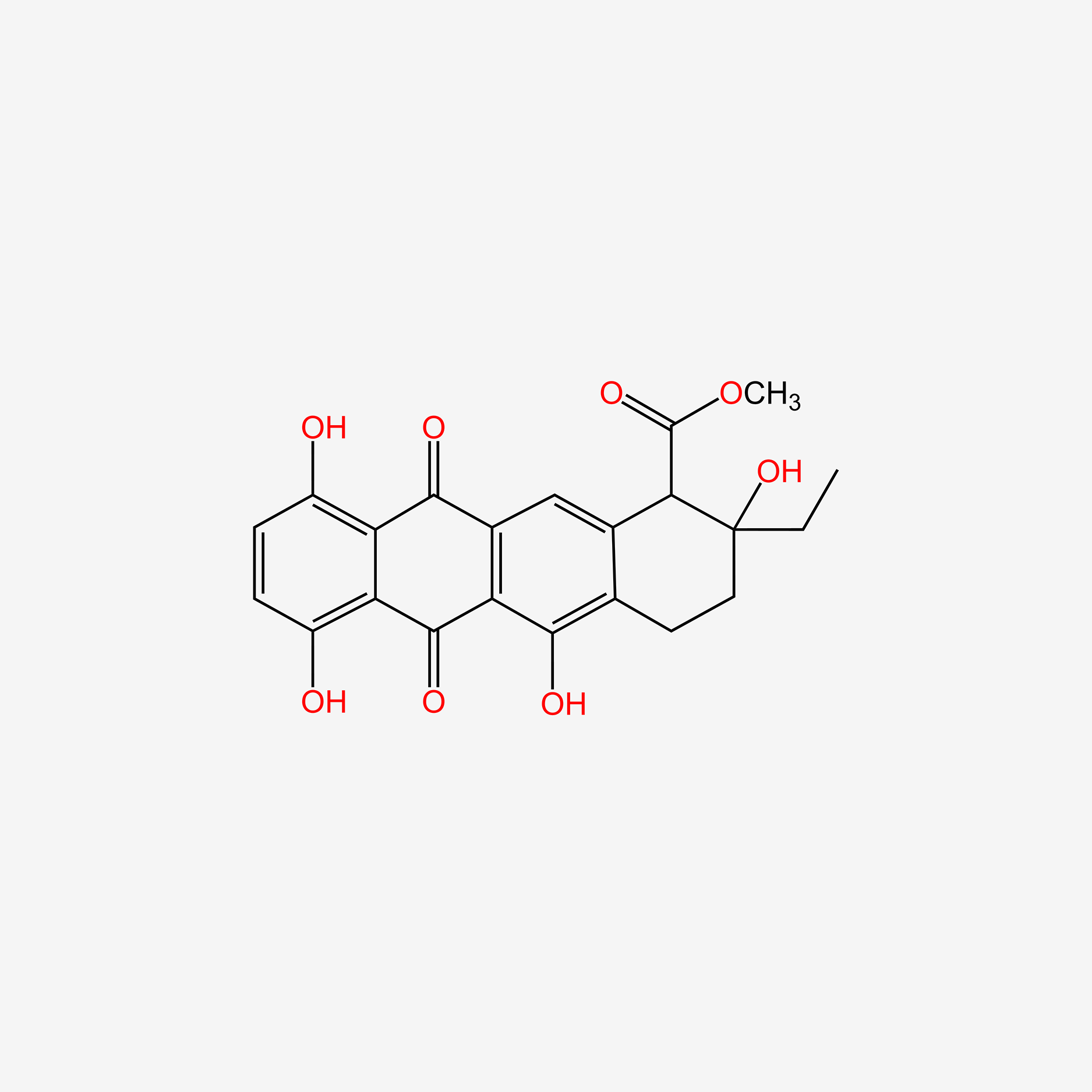 |
0.343 | D07MOX | 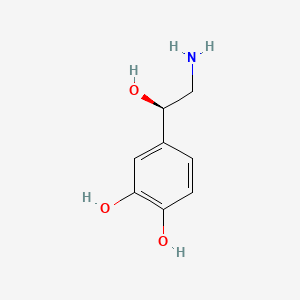 |
0.294 | ||
| ENC000087 | 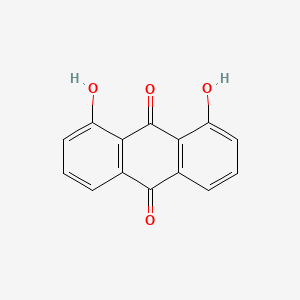 |
0.342 | D0R3JB | 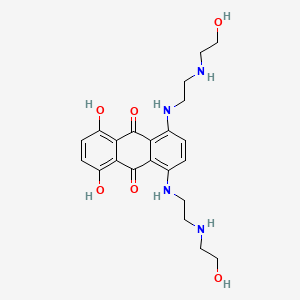 |
0.281 | ||
| ENC004888 | 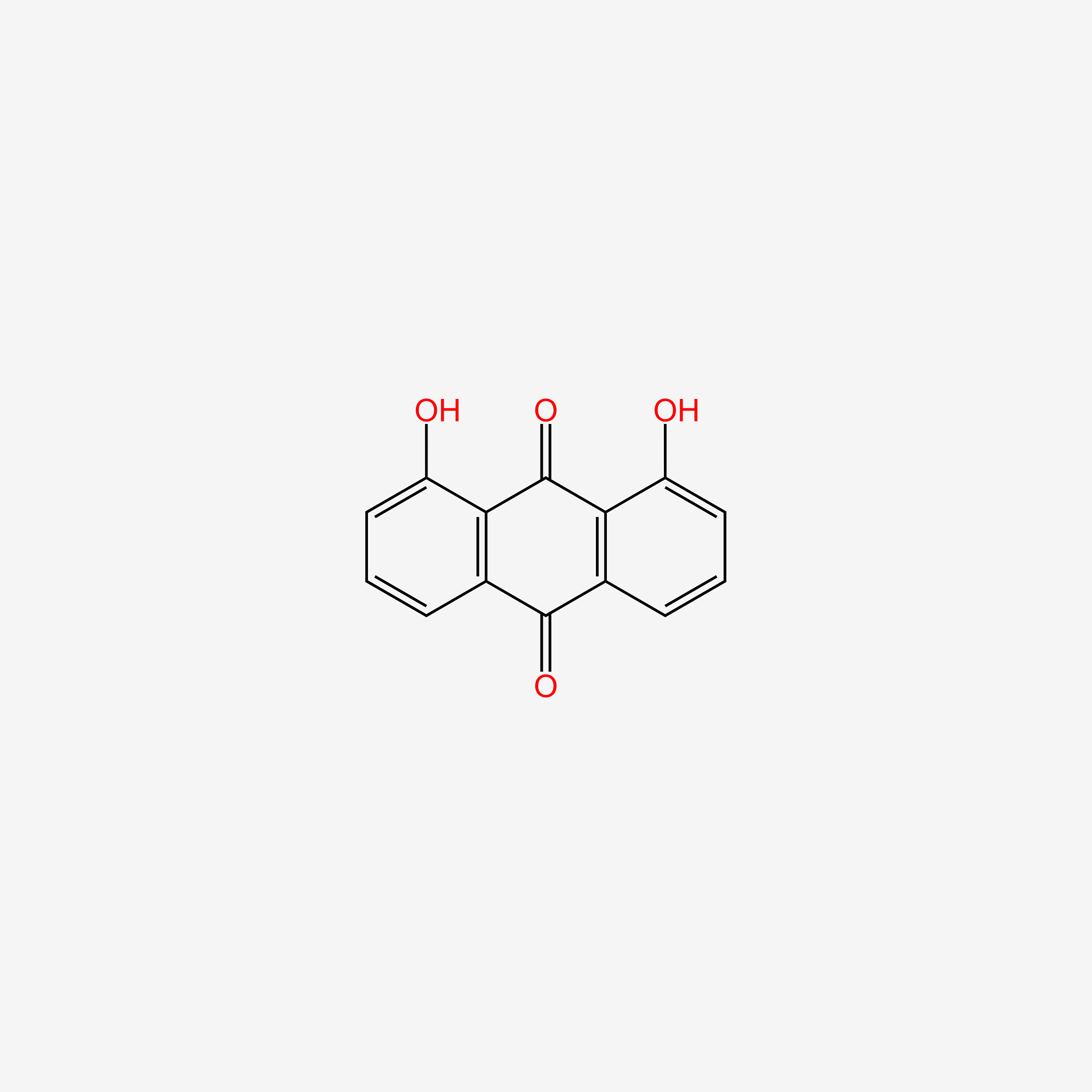 |
0.342 | D08HVR | 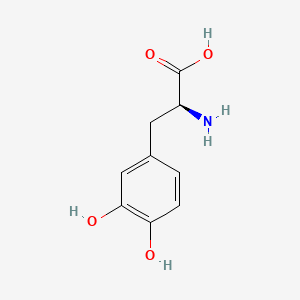 |
0.274 | ||
| ENC005342 | 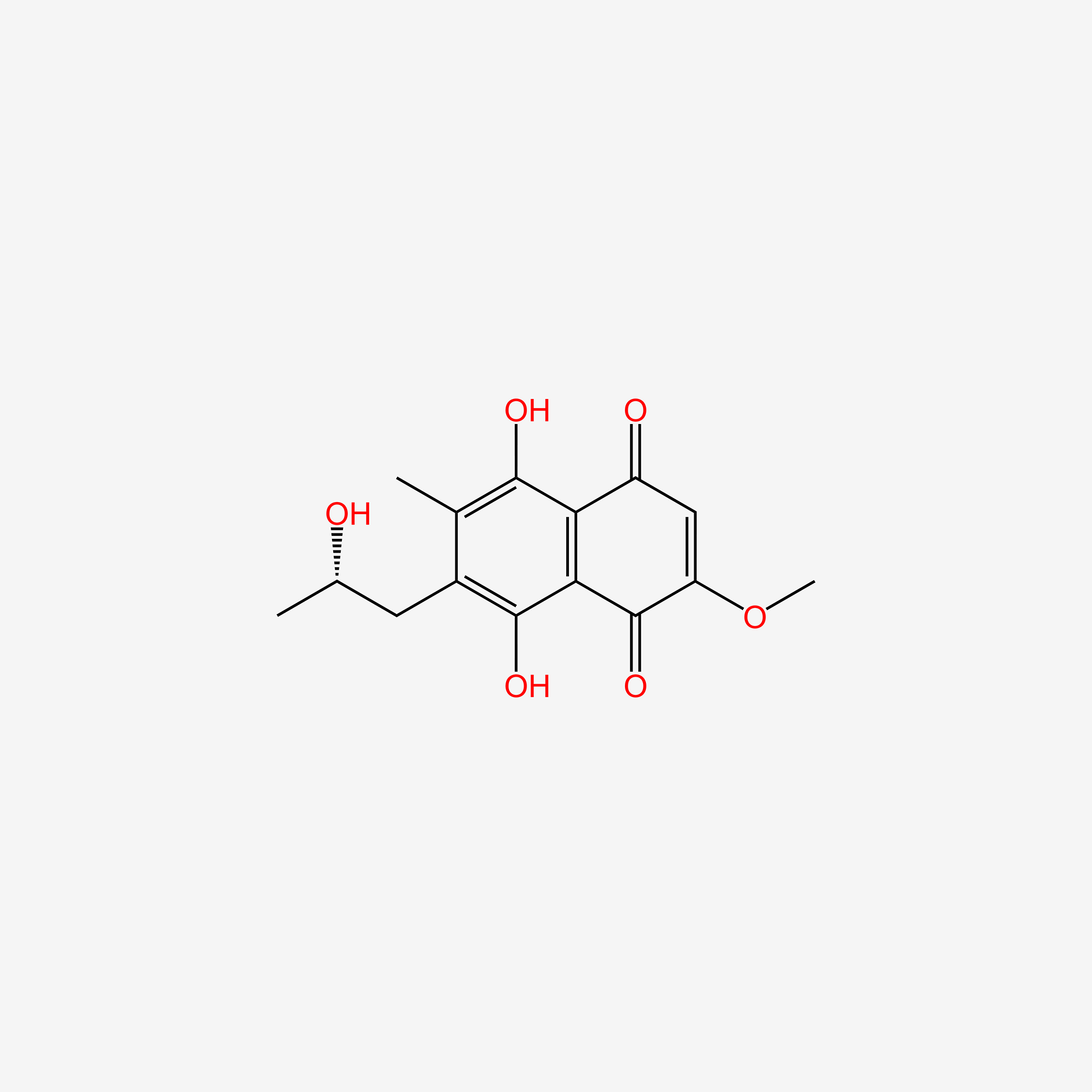 |
0.337 | D0BA6T | 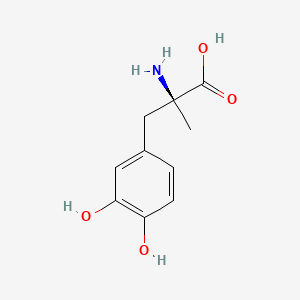 |
0.267 | ||
| ENC000929 | 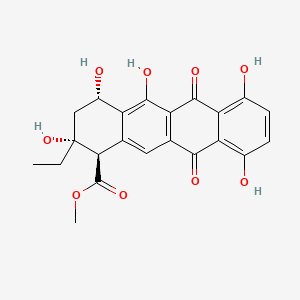 |
0.337 | D0I3RO | 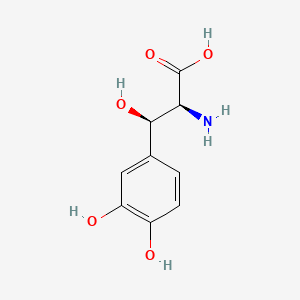 |
0.267 | ||
| ENC000337 | 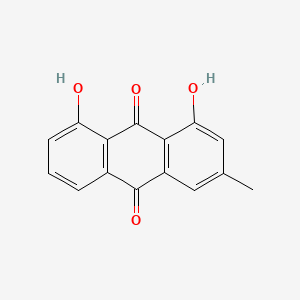 |
0.333 | D0V9EN | 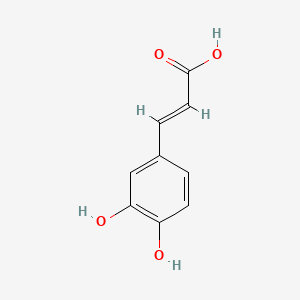 |
0.264 | ||
| ENC000934 | 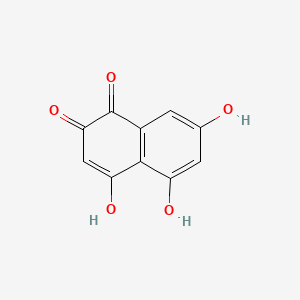 |
0.333 | D0U0OT | 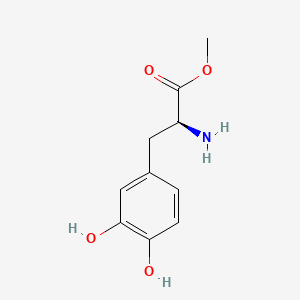 |
0.263 | ||