NPs Basic Information
|
Name |
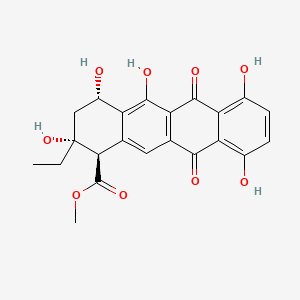
Epsilon-pyrromycinone
|
| Molecular Formula | C22H20O9 | |
| IUPAC Name* |
methyl (1R,2R,4S)-2-ethyl-2,4,5,7,10-pentahydroxy-6,11-dioxo-3,4-dihydro-1H-tetracene-1-carboxylate
|
|
| SMILES |
CC[C@]1(C[C@@H](C2=C(C3=C(C=C2[C@H]1C(=O)OC)C(=O)C4=C(C=CC(=C4C3=O)O)O)O)O)O
|
|
| InChI |
InChI=1S/C22H20O9/c1-3-22(30)7-12(25)13-8(17(22)21(29)31-2)6-9-14(19(13)27)20(28)16-11(24)5-4-10(23)15(16)18(9)26/h4-6,12,17,23-25,27,30H,3,7H2,1-2H3/t12-,17-,22+/m0/s1
|
|
| InChIKey |
RWCVSDKDFSVZNF-KRYGIPSASA-N
|
|
| Synonyms |
Rutilantinone; Epsilon-pyrromycinone; 21288-61-9; Eta-Pyrromycinon; Rutilantinon; Antibiotic MA 144D2; PYRROMYCINONE, EPSILON; 1-hydroxyaklavinone; Galirubine; MA 144D2; epsilon-pyrromy-cinone; NSC 114778; Spectrum_000795; Spectrum2_000452; Spectrum3_001155; Spectrum4_001432; Spectrum5_000284; BSPBio_002709; KBioGR_001904; KBioSS_001275; SPECTRUM201606; SPBio_000524; CHEMBL3039086; KBio2_001275; KBio2_003843; KBio2_006411; KBio3_002209; CHEBI:108590; HMS1923A07; CCG-38694; SDCCGMLS-0066382.P001; NCGC00178516-01; 1-Naphthacenecarboxylic acid, 1,2,3,4,6,11-hexahydro-2-ethyl-2,4,5,7,10-pentahydroxy-6,11-dioxo-, methyl ester, (1R,2R,4S)-; 1-Naphthacenecarboxylic acid, 2-ethyl-1,2,3,4,6,11-hexahydro-2,4,5,7,10-pentahydroxy-6,11-dioxo-, methyl ester, (1R,2R,4S)-; SR-05000002666; SR-05000002666-1; BRD-K11258719-001-03-2; Q27187513; (1R,2R,4S)-2-ethyl-2,4,5,7,10-pentahydroxy-6,11-dioxo-3,4-dihydro-1H-tetracene-1-carboxylic acid methyl ester
|
|
| CAS | 21288-61-9 | |
| PubChem CID | 159908 | |
| ChEMBL ID | CHEMBL3039086 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 428.4 | ALogp: | 2.7 |
| HBD: | 5 | HBA: | 9 |
| Rotatable Bonds: | 3 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 162.0 | Aromatic Rings: | 4 |
| Heavy Atoms: | 31 | QED Weighted: | 0.303 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.691 | MDCK Permeability: | 0.00000511 |
| Pgp-inhibitor: | 0.024 | Pgp-substrate: | 0.046 |
| Human Intestinal Absorption (HIA): | 0.964 | 20% Bioavailability (F20%): | 0.024 |
| 30% Bioavailability (F30%): | 0.978 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.004 | Plasma Protein Binding (PPB): | 95.16% |
| Volume Distribution (VD): | 1.071 | Fu: | 9.36% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.638 | CYP1A2-substrate: | 0.878 |
| CYP2C19-inhibitor: | 0.025 | CYP2C19-substrate: | 0.054 |
| CYP2C9-inhibitor: | 0.55 | CYP2C9-substrate: | 0.577 |
| CYP2D6-inhibitor: | 0.023 | CYP2D6-substrate: | 0.164 |
| CYP3A4-inhibitor: | 0.106 | CYP3A4-substrate: | 0.07 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 5.467 | Half-life (T1/2): | 0.334 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.004 | Human Hepatotoxicity (H-HT): | 0.078 |
| Drug-inuced Liver Injury (DILI): | 0.952 | AMES Toxicity: | 0.71 |
| Rat Oral Acute Toxicity: | 0.039 | Maximum Recommended Daily Dose: | 0.107 |
| Skin Sensitization: | 0.693 | Carcinogencity: | 0.042 |
| Eye Corrosion: | 0.003 | Eye Irritation: | 0.767 |
| Respiratory Toxicity: | 0.072 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC005576 | 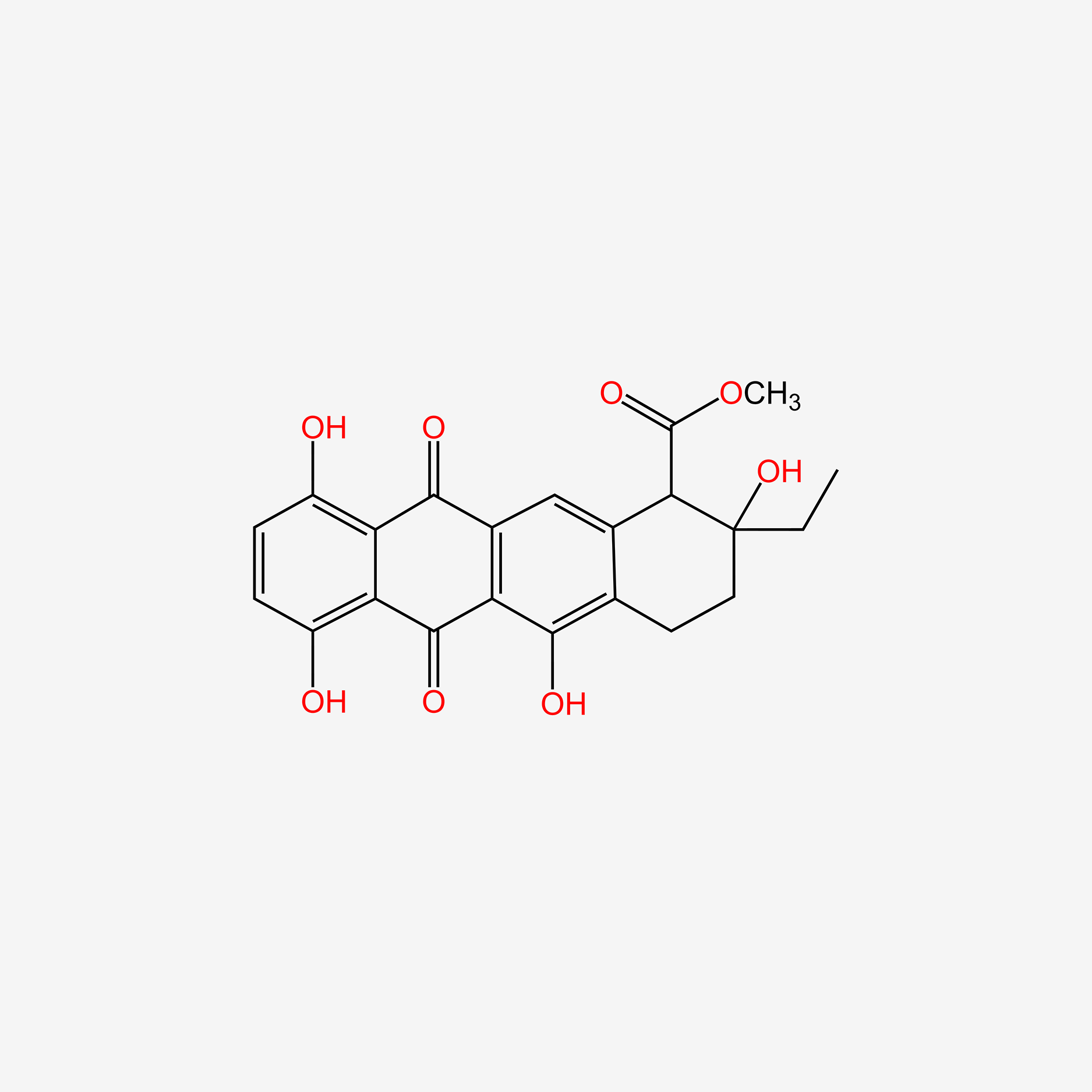 |
0.763 | D01UBX | 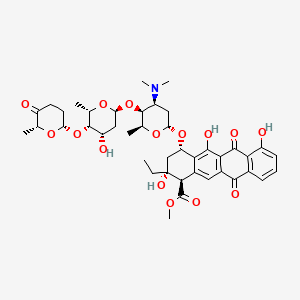 |
0.399 | ||
| ENC003228 | 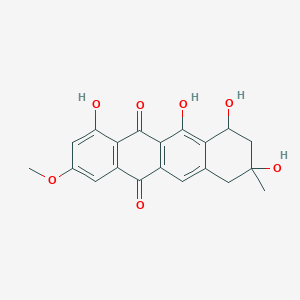 |
0.458 | D0C9XJ | 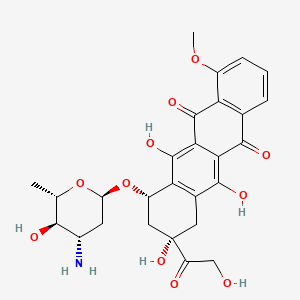 |
0.390 | ||
| ENC000336 | 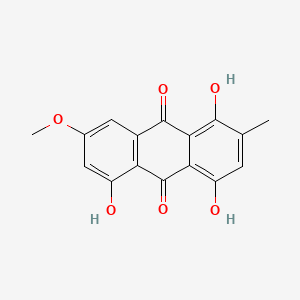 |
0.420 | D07VLY | 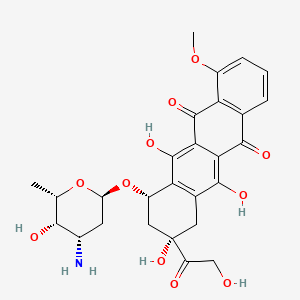 |
0.390 | ||
| ENC000335 | 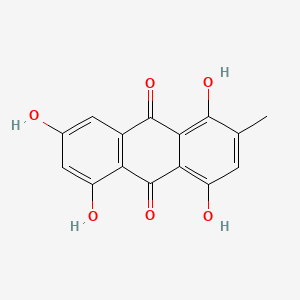 |
0.404 | D01XWG |  |
0.378 | ||
| ENC005279 |  |
0.402 | D01XDL |  |
0.351 | ||
| ENC002067 |  |
0.394 | D0T5XN |  |
0.338 | ||
| ENC002125 |  |
0.392 | D0R9WP |  |
0.336 | ||
| ENC004746 |  |
0.389 | D07JHH |  |
0.331 | ||
| ENC000935 | 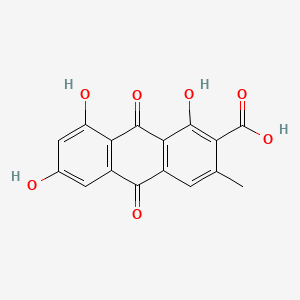 |
0.385 | D0H1AR | 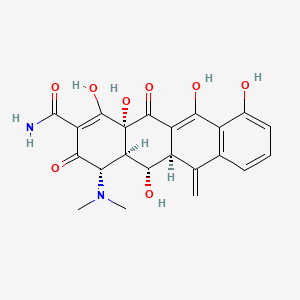 |
0.325 | ||
| ENC002767 | 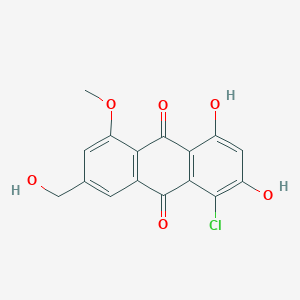 |
0.381 | D0T8EH | 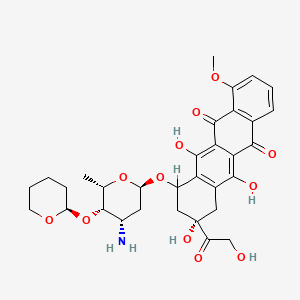 |
0.325 | ||