NPs Basic Information
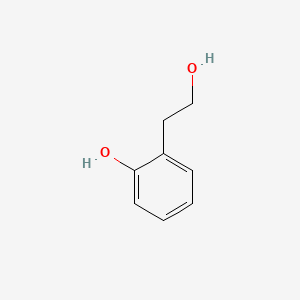
|
Name |
2-Hydroxyphenethyl alcohol
|
| Molecular Formula | C8H10O2 | |
| IUPAC Name* |
2-(2-hydroxyethyl)phenol
|
|
| SMILES |
C1=CC=C(C(=C1)CCO)O
|
|
| InChI |
InChI=1S/C8H10O2/c9-6-5-7-3-1-2-4-8(7)10/h1-4,9-10H,5-6H2
|
|
| InChIKey |
ABFCOJLLBHXNOU-UHFFFAOYSA-N
|
|
| Synonyms |
2-(2-Hydroxyethyl)phenol; 2-Hydroxyphenethyl alcohol; 7768-28-7; 2-Hydroxyphenylethanol; 2-(2-Hydroxyphenyl)ethanol; Benzeneethanol, 2-hydroxy-; 2-Hydroxybenzeneethanol; o-(2-Hydroxyethyl)phenol; o-Hydroxyphenethyl alcohol; 2-(o-Hydroxyphenyl)ethanol; 2-hydroxy-benzeneethanol; NSC 101845; C6AV79GN9F; beta-(o-Hydroxyphenyl)ethanol; MFCD00002890; NSC-101845; 2-phenolethanol; EINECS 231-863-3; 2-Hydroxyethylphenol; UNII-C6AV79GN9F; SCHEMBL43837; 2-(2-hydroxy-ethyl)-phenol; 2-(2-hydroxyphenyl)-ethanol; Phenethyl alcohol, o-hydroxy-; CHEMBL4075005; .beta.-(o-Hydroxyphenyl)ethanol; CHEBI:64803; DTXSID90228312; 2-Hydroxyphenethyl alcohol, 99%; 2-(2-Hydroxyethyl)phenol, 95%; ZINC406927; ACT04258; AMY37713; NSC101845; AKOS005259862; AB17761; CS-W016503; AS-10136; SY066615; DB-075363; FT-0648868; EN300-179020; 768H287; W-104304; Q27133443
|
|
| CAS | 7768-28-7 | |
| PubChem CID | 82200 | |
| ChEMBL ID | CHEMBL4075005 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 138.16 | ALogp: | 1.2 |
| HBD: | 2 | HBA: | 2 |
| Rotatable Bonds: | 2 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 40.5 | Aromatic Rings: | 1 |
| Heavy Atoms: | 10 | QED Weighted: | 0.647 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.28 | MDCK Permeability: | 0.00002080 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.002 |
| Human Intestinal Absorption (HIA): | 0.154 | 20% Bioavailability (F20%): | 0.977 |
| 30% Bioavailability (F30%): | 0.992 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.151 | Plasma Protein Binding (PPB): | 38.00% |
| Volume Distribution (VD): | 2.63 | Fu: | 46.33% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.565 | CYP1A2-substrate: | 0.553 |
| CYP2C19-inhibitor: | 0.117 | CYP2C19-substrate: | 0.145 |
| CYP2C9-inhibitor: | 0.024 | CYP2C9-substrate: | 0.5 |
| CYP2D6-inhibitor: | 0.081 | CYP2D6-substrate: | 0.633 |
| CYP3A4-inhibitor: | 0.018 | CYP3A4-substrate: | 0.247 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 13 | Half-life (T1/2): | 0.919 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.014 | Human Hepatotoxicity (H-HT): | 0.037 |
| Drug-inuced Liver Injury (DILI): | 0.053 | AMES Toxicity: | 0.222 |
| Rat Oral Acute Toxicity: | 0.395 | Maximum Recommended Daily Dose: | 0.011 |
| Skin Sensitization: | 0.854 | Carcinogencity: | 0.502 |
| Eye Corrosion: | 0.947 | Eye Irritation: | 0.993 |
| Respiratory Toxicity: | 0.042 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC005498 | 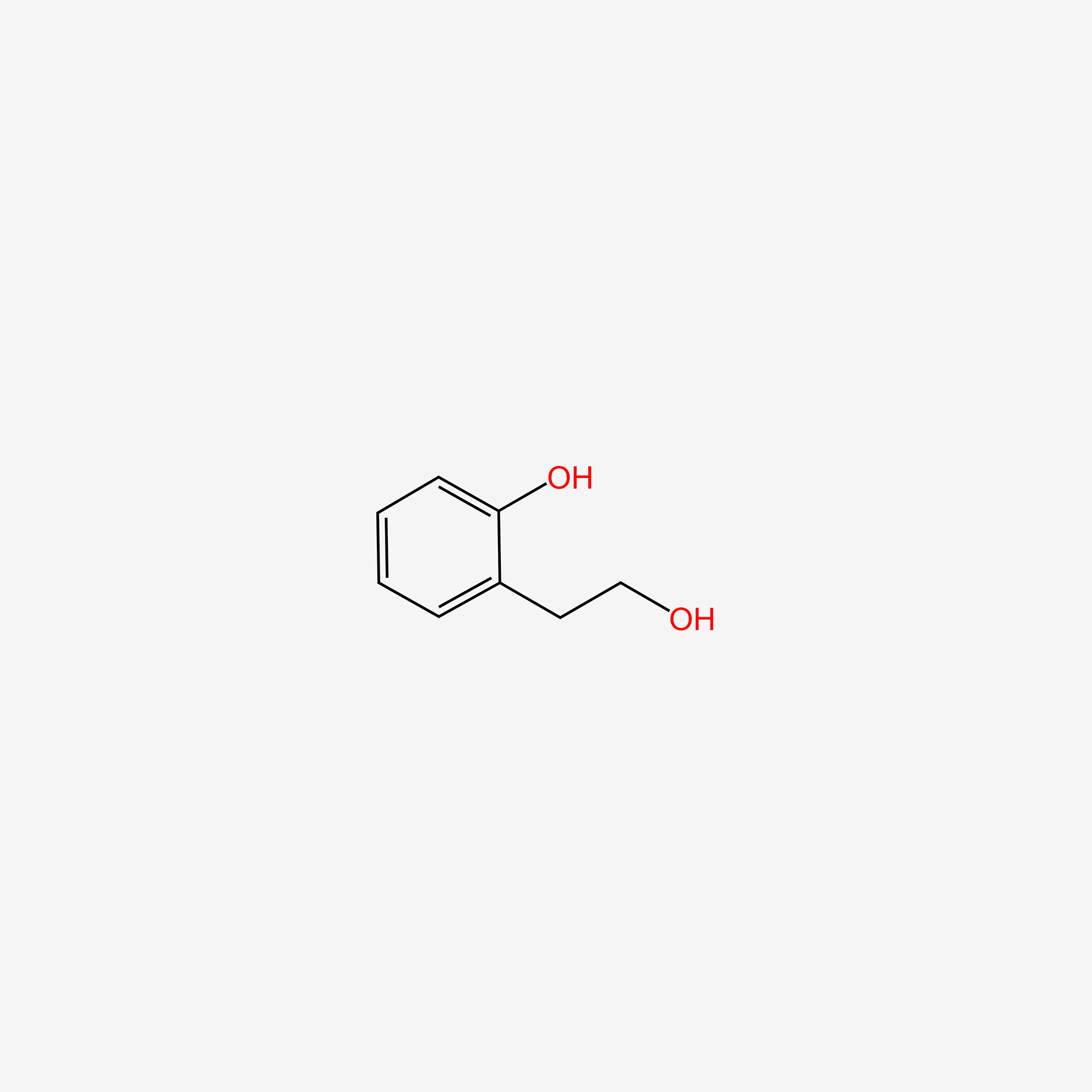 |
1.000 | D07HBX |  |
0.447 | ||
| ENC000409 | 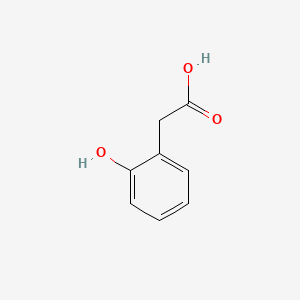 |
0.568 | D05OIS | 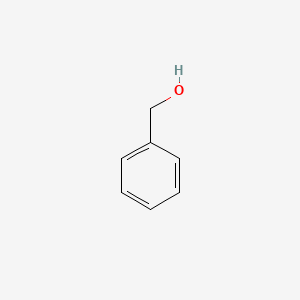 |
0.378 | ||
| ENC000021 | 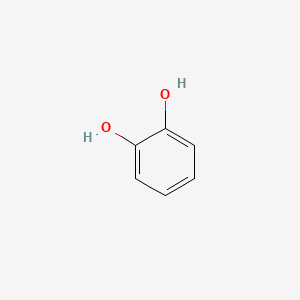 |
0.515 | D0A5CM | 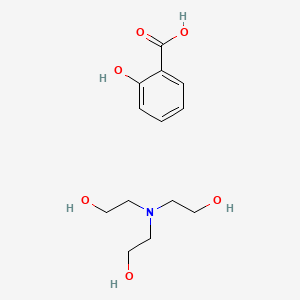 |
0.344 | ||
| ENC000128 | 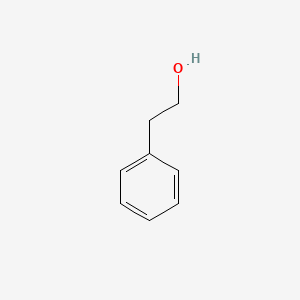 |
0.500 | D02YYF | 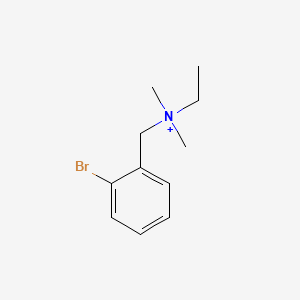 |
0.340 | ||
| ENC000363 | 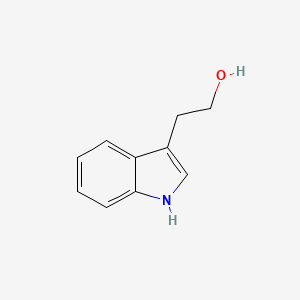 |
0.500 | D0P9AC | 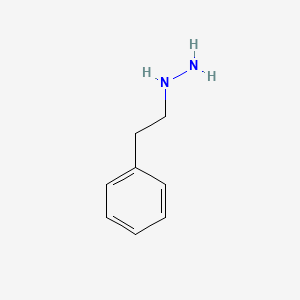 |
0.326 | ||
| ENC000756 | 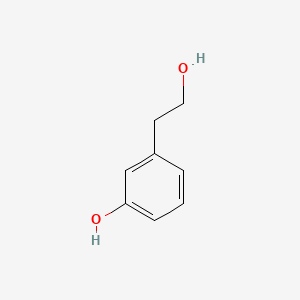 |
0.474 | D0F5ZM | 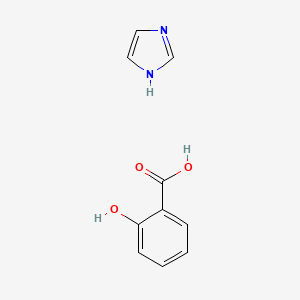 |
0.321 | ||
| ENC000028 |  |
0.471 | D0T7OW |  |
0.318 | ||
| ENC003028 |  |
0.452 | D06BQU |  |
0.297 | ||
| ENC001315 | 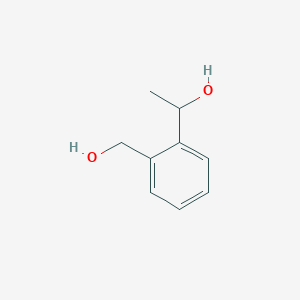 |
0.450 | D0P2GK | 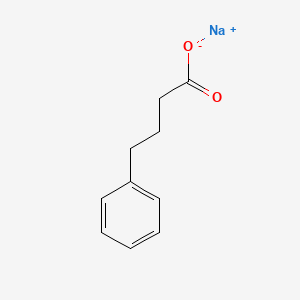 |
0.286 | ||
| ENC000350 | 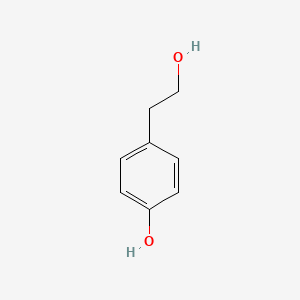 |
0.436 | D03UOT | 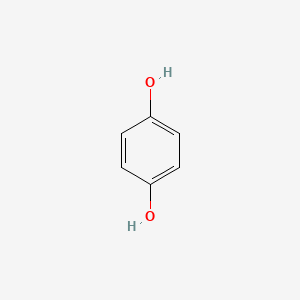 |
0.282 | ||