NPs Basic Information
|
Name |
Vinyl 2-ethylhexanoate
|
| Molecular Formula | C10H18O2 | |
| IUPAC Name* |
ethenyl 2-ethylhexanoate
|
|
| SMILES |
CCCCC(CC)C(=O)OC=C
|
|
| InChI |
InChI=1S/C10H18O2/c1-4-7-8-9(5-2)10(11)12-6-3/h6,9H,3-5,7-8H2,1-2H3
|
|
| InChIKey |
IGBZOHMCHDADGY-UHFFFAOYSA-N
|
|
| Synonyms |
VINYL 2-ETHYLHEXANOATE; 2-Ethylhexanoic Acid Vinyl Ester; 94-04-2; ethenyl 2-ethylhexanoate; Vinyl 2-ethylhexoate; 2-Ethylhexanoic acid, vinyl ester; Hexanoic acid, 2-ethyl-, ethenyl ester; Vynate 2EH; 2-Ethylhexoic acid, vinyl ester; Vinyl-2-ethylhexanoate; Hexanoic acid, 2-ethyl-, vinyl ester; Vinylester kyseliny 2-ethylkapronove; 1QLD4A946M; NSC-5312; Vinyl-2-ethylhexoate; NSC 5312; EINECS 202-297-4; BRN 1767220; UNII-1QLD4A946M; AI3-24890; Vinylester kyseliny 2-ethylkapronove [Czech]; vinyl2-ethylhexanoate; VEOVA EH; 2-Ethylhexanoic acid vinyl; EC 202-297-4; SCHEMBL35266; WLN: 4Y2&VO1U1; DTXSID4052633; IGBZOHMCHDADGY-UHFFFAOYSA-; NSC5312; 2-ethyl hexanoic acid vinyl ester; MFCD00009488; AKOS015836289; BS-23881; 2-ETHYL-HEXANOIC ACID, VINYL ESTER; E0404; FT-0694071; Vinyl 2-Ethylhexanoate (stabilized with MEHQ); Q27252762
|
|
| CAS | 94-04-2 | |
| PubChem CID | 62343 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 170.25 | ALogp: | 3.5 |
| HBD: | 0 | HBA: | 2 |
| Rotatable Bonds: | 7 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 26.3 | Aromatic Rings: | 0 |
| Heavy Atoms: | 12 | QED Weighted: | 0.449 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.424 | MDCK Permeability: | 0.00002610 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.004 | 20% Bioavailability (F20%): | 0.003 |
| 30% Bioavailability (F30%): | 0.383 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.995 | Plasma Protein Binding (PPB): | 39.26% |
| Volume Distribution (VD): | 1.021 | Fu: | 71.71% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.825 | CYP1A2-substrate: | 0.466 |
| CYP2C19-inhibitor: | 0.551 | CYP2C19-substrate: | 0.815 |
| CYP2C9-inhibitor: | 0.387 | CYP2C9-substrate: | 0.544 |
| CYP2D6-inhibitor: | 0.012 | CYP2D6-substrate: | 0.295 |
| CYP3A4-inhibitor: | 0.225 | CYP3A4-substrate: | 0.375 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 6.449 | Half-life (T1/2): | 0.658 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.01 | Human Hepatotoxicity (H-HT): | 0.057 |
| Drug-inuced Liver Injury (DILI): | 0.04 | AMES Toxicity: | 0.005 |
| Rat Oral Acute Toxicity: | 0.027 | Maximum Recommended Daily Dose: | 0.187 |
| Skin Sensitization: | 0.948 | Carcinogencity: | 0.883 |
| Eye Corrosion: | 0.967 | Eye Irritation: | 0.971 |
| Respiratory Toxicity: | 0.86 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
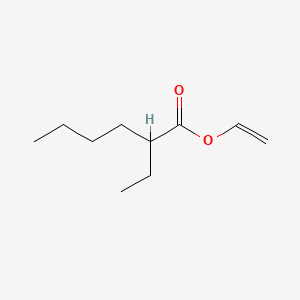
| ENC000833 | 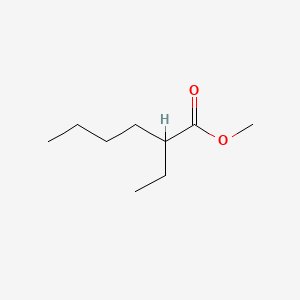 |
0.649 | D0Y3KG | 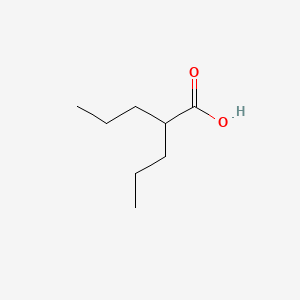 |
0.349 | ||
| ENC000306 | 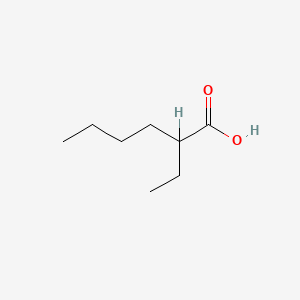 |
0.568 | D01QLH | 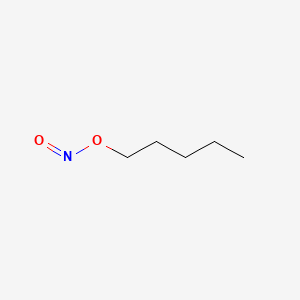 |
0.256 | ||
| ENC000212 | 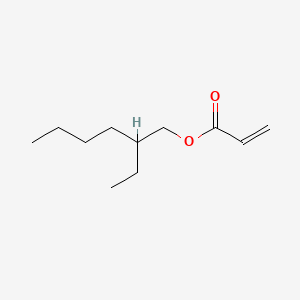 |
0.489 | D07CNL | 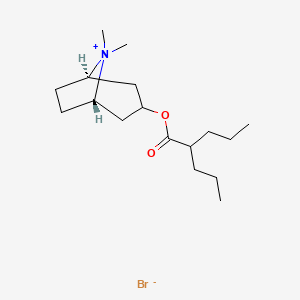 |
0.239 | ||
| ENC002444 | 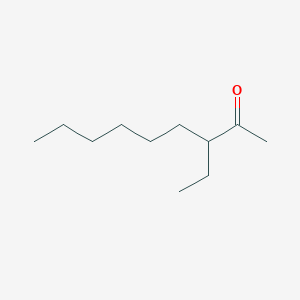 |
0.455 | D0ZK8H | 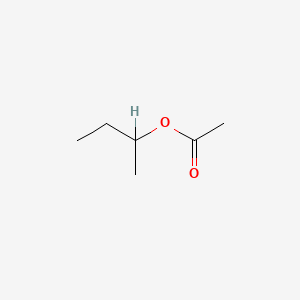 |
0.238 | ||
| ENC000211 | 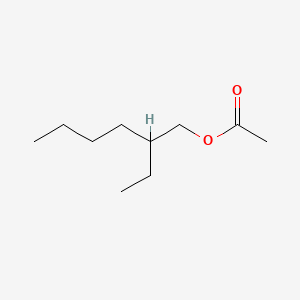 |
0.422 | D0X4FM | 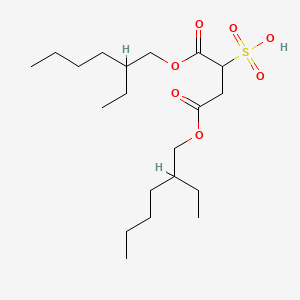 |
0.230 | ||
| ENC000512 | 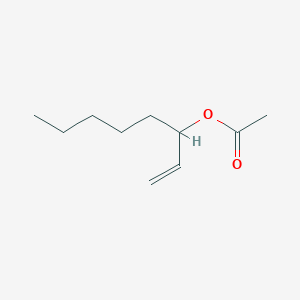 |
0.422 | D0R3QY |  |
0.222 | ||
| ENC001211 | 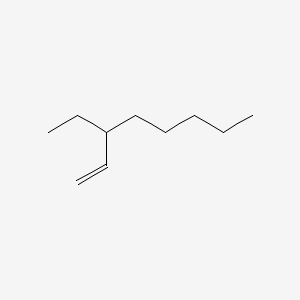 |
0.405 | D0CT4D | 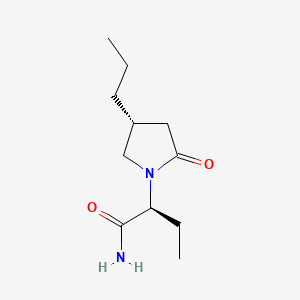 |
0.220 | ||
| ENC000849 | 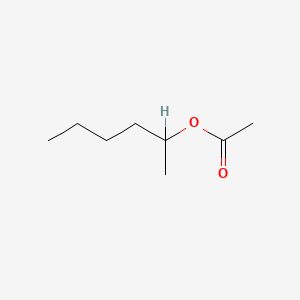 |
0.381 | D03LGY | 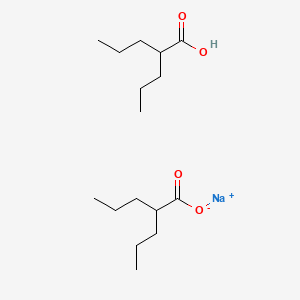 |
0.214 | ||
| ENC000747 | 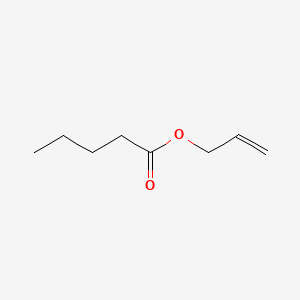 |
0.372 | D08EVN | 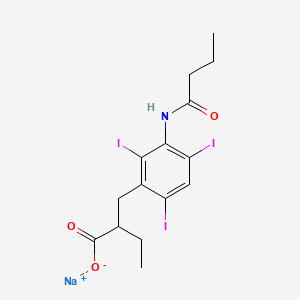 |
0.213 | ||
| ENC000570 | 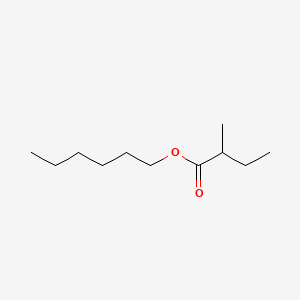 |
0.367 | D0AY9Q | 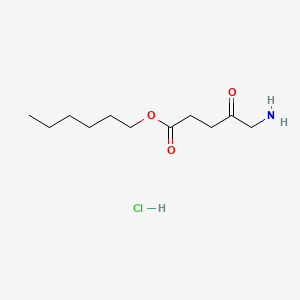 |
0.213 | ||