NPs Basic Information
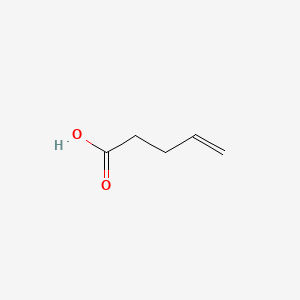
|
Name |
4-Pentenoic acid
|
| Molecular Formula | C5H8O2 | |
| IUPAC Name* |
pent-4-enoic acid
|
|
| SMILES |
C=CCCC(=O)O
|
|
| InChI |
InChI=1S/C5H8O2/c1-2-3-4-5(6)7/h2H,1,3-4H2,(H,6,7)
|
|
| InChIKey |
HVAMZGADVCBITI-UHFFFAOYSA-N
|
|
| Synonyms |
4-PENTENOIC ACID; Pent-4-enoic acid; 591-80-0; Allylacetic acid; Allyl acetic acid; 3-Vinylpropionic acid; 4 PA; FEMA No. 2843; .DELTA.4-Pentenoic acid; D4S77Y29FB; CHEBI:35936; C5:1n-1; NSC-9000; NSC-20944; WLN: QV3U1; delta 4-Pentenoic acid; NSC 9000; EINECS 209-732-7; NSC 20944; BRN 1633696; UNII-D4S77Y29FB; Allylessigsaeure; 4-Pentensaeure; 4-pentenic acid; Pent-4-enoicacid; MFCD00004408; delta4-Pentenoic acid; 4-penten-1-oic acid; Delta(4)-pentenoic acid; 4-Pentenoic acid, 97%; 3-Butene-1-carboxylic acid; DSSTox_CID_24448; DSSTox_RID_80236; DSSTox_GSID_44448; 4-02-00-01542 (Beilstein Handbook Reference); SCHEMBL115342; CHEMBL3185583; DTXSID0044448; SCHEMBL13341412; FEMA 2843; 4-PENTENOIC ACID [FHFI]; NSC9000; ACT03062; AMY12553; HY-Y0624; NSC20944; ZINC1648357; Tox21_302069; BBL027458; LMFA01030007; s6268; STL280305; AKOS006221010; CS-W009138; NCGC00188966-01; NCGC00255889-01; AS-11763; CAS-591-80-0; 4-Pentenoic acid, >=98%, stabilized, FG; FT-0676299; P0645; EN300-64825; O12040; J-519555; Q27116641; F2190-0007; Z995028722
|
|
| CAS | 591-80-0 | |
| PubChem CID | 61138 | |
| ChEMBL ID | CHEMBL3185583 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 100.12 | ALogp: | 0.8 |
| HBD: | 1 | HBA: | 2 |
| Rotatable Bonds: | 3 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 37.3 | Aromatic Rings: | 0 |
| Heavy Atoms: | 7 | QED Weighted: | 0.545 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.935 | MDCK Permeability: | 0.00006050 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0 |
| Human Intestinal Absorption (HIA): | 0.006 | 20% Bioavailability (F20%): | 0.256 |
| 30% Bioavailability (F30%): | 0.082 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.57 | Plasma Protein Binding (PPB): | 69.54% |
| Volume Distribution (VD): | 0.23 | Fu: | 25.84% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.016 | CYP1A2-substrate: | 0.11 |
| CYP2C19-inhibitor: | 0.026 | CYP2C19-substrate: | 0.062 |
| CYP2C9-inhibitor: | 0.011 | CYP2C9-substrate: | 0.908 |
| CYP2D6-inhibitor: | 0.008 | CYP2D6-substrate: | 0.27 |
| CYP3A4-inhibitor: | 0.01 | CYP3A4-substrate: | 0.055 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 6.676 | Half-life (T1/2): | 0.806 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.007 | Human Hepatotoxicity (H-HT): | 0.123 |
| Drug-inuced Liver Injury (DILI): | 0.018 | AMES Toxicity: | 0.02 |
| Rat Oral Acute Toxicity: | 0.85 | Maximum Recommended Daily Dose: | 0.025 |
| Skin Sensitization: | 0.315 | Carcinogencity: | 0.096 |
| Eye Corrosion: | 0.987 | Eye Irritation: | 0.992 |
| Respiratory Toxicity: | 0.385 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000647 | 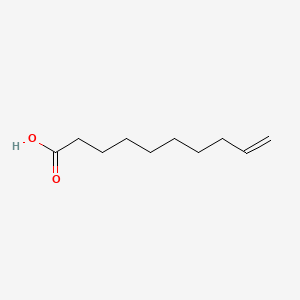 |
0.457 | D0R3QY |  |
0.464 | ||
| ENC000677 | 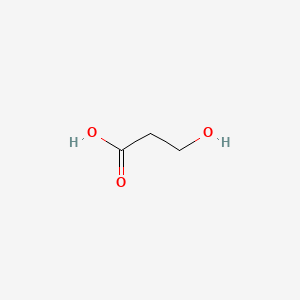 |
0.435 | D0Z5BC | 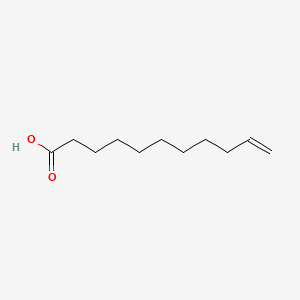 |
0.421 | ||
| ENC000018 | 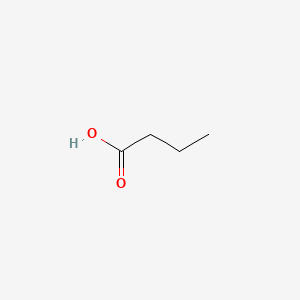 |
0.435 | D0EP8X | 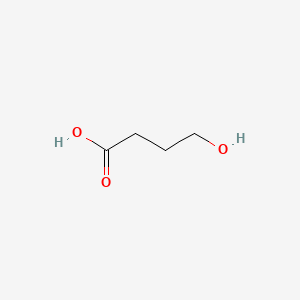 |
0.385 | ||
| ENC000686 | 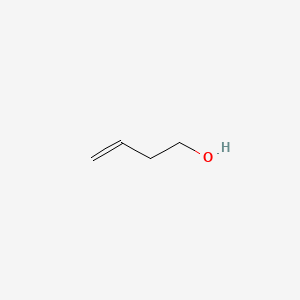 |
0.409 | D06VNK | 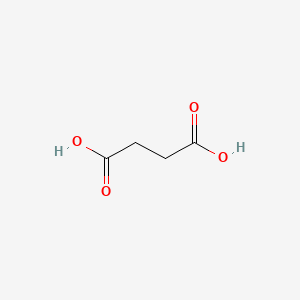 |
0.357 | ||
| ENC000148 | 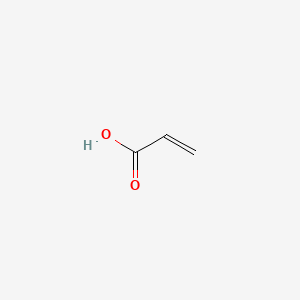 |
0.381 | D0Y7ZD | 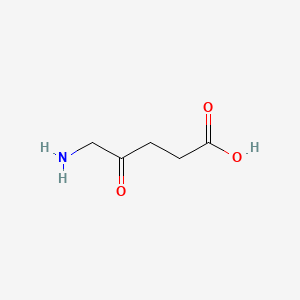 |
0.323 | ||
| ENC000445 | 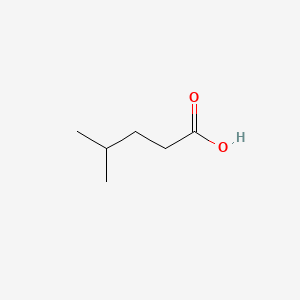 |
0.357 | D0M8AB | 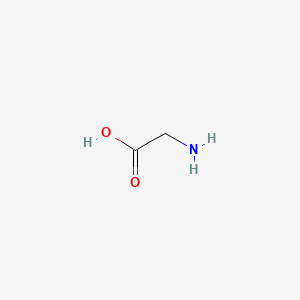 |
0.318 | ||
| ENC000062 | 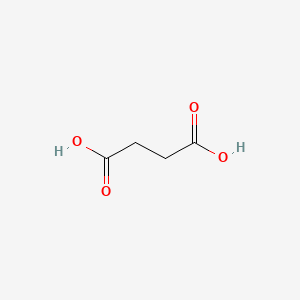 |
0.357 | D0O4GY | 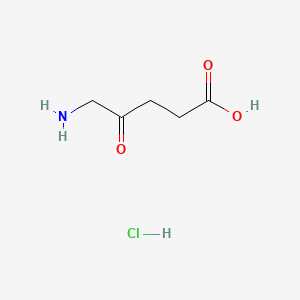 |
0.313 | ||
| ENC000070 | 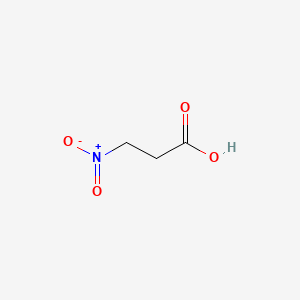 |
0.357 | D0FD0H | 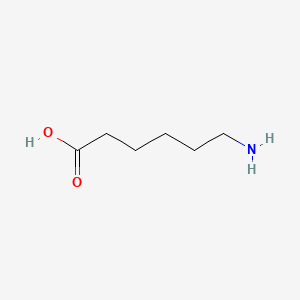 |
0.313 | ||
| ENC000643 | 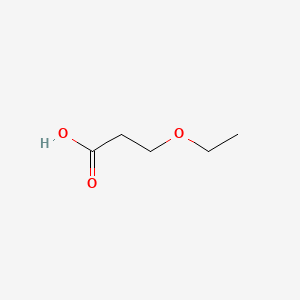 |
0.345 | D00ENY | 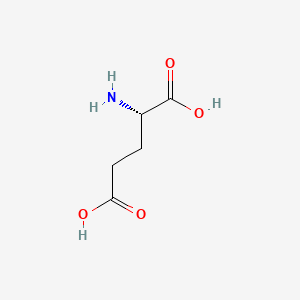 |
0.303 | ||
| ENC000315 | 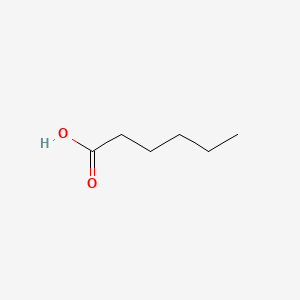 |
0.345 | D0C6OQ | 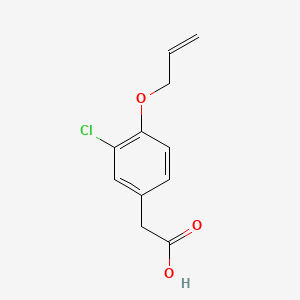 |
0.255 | ||