NPs Basic Information
|
Name |
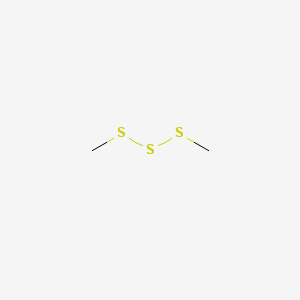
Dimethyl trisulfide
|
| Molecular Formula | C2H6S3 | |
| IUPAC Name* |
(methyltrisulfanyl)methane
|
|
| SMILES |
CSSSC
|
|
| InChI |
InChI=1S/C2H6S3/c1-3-5-4-2/h1-2H3
|
|
| InChIKey |
YWHLKYXPLRWGSE-UHFFFAOYSA-N
|
|
| Synonyms |
Dimethyl trisulfide; 3658-80-8; Trisulfide, dimethyl; Methyl trisulfide; 2,3,4-Trithiapentane; Dimethyl trisulphide; (methyltrisulfanyl)methane; DIMETHYLTRISULFIDE; dimethyltrisulfane; DMTS; FEMA No. 3275; CH3SSSCH3; 3E691T3NL1; NSC-97324; UNII-3E691T3NL1; Dimethyl trisufide; EINECS 222-910-9; 2,4-Trithiapentane; NSC 97324; trisulfane, dimethyl-; 1,3-Dimethyltrisulfane; AI3-26172; Dimethyl Trisulfide-[d6]; 1,3-Dimethyltrisulfane #; SCHEMBL446658; methylsulfanyldisulfanyl-methane; CHEBI:4614; DTXSID9063118; DIMETHYL TRISULFIDE [FHFI]; Dimethyl trisulfide, >=98%, FG; NSC97324; MFCD00039808; NSC801680; s6311; AKOS015897465; NSC-801680; Dimethyl trisulfide, analytical standard; BS-43830; 1,3-Dimethyltrisulfane (ACD/Name 4.0); DB-003633; HY-128454; CS-0099182; D3418; FT-0625104; FT-0667568; C08372; D90187; 658D808; A823301; Q-100435; Q5277321; FLAMMABLE LIQUID, N.O.S. (DIMETHYL TRISULPHIDE)
|
|
| CAS | 3658-80-8 | |
| PubChem CID | 19310 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 126.3 | ALogp: | 1.3 |
| HBD: | 0 | HBA: | 3 |
| Rotatable Bonds: | 2 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 75.9 | Aromatic Rings: | 0 |
| Heavy Atoms: | 5 | QED Weighted: | 0.522 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.558 | MDCK Permeability: | 0.00002210 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.004 |
| Human Intestinal Absorption (HIA): | 0.015 | 20% Bioavailability (F20%): | 0.003 |
| 30% Bioavailability (F30%): | 0.685 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.531 | Plasma Protein Binding (PPB): | 34.11% |
| Volume Distribution (VD): | 1.063 | Fu: | 59.00% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.921 | CYP1A2-substrate: | 0.92 |
| CYP2C19-inhibitor: | 0.912 | CYP2C19-substrate: | 0.869 |
| CYP2C9-inhibitor: | 0.718 | CYP2C9-substrate: | 0.657 |
| CYP2D6-inhibitor: | 0.022 | CYP2D6-substrate: | 0.671 |
| CYP3A4-inhibitor: | 0.009 | CYP3A4-substrate: | 0.314 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 11.516 | Half-life (T1/2): | 0.315 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.148 | Human Hepatotoxicity (H-HT): | 0.126 |
| Drug-inuced Liver Injury (DILI): | 0.157 | AMES Toxicity: | 0.211 |
| Rat Oral Acute Toxicity: | 0.965 | Maximum Recommended Daily Dose: | 0.111 |
| Skin Sensitization: | 0.95 | Carcinogencity: | 0.229 |
| Eye Corrosion: | 0.984 | Eye Irritation: | 0.996 |
| Respiratory Toxicity: | 0.986 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000743 |  |
0.706 | D08HVE |  |
0.105 | ||
| ENC000417 |  |
0.571 | D0U3IG | 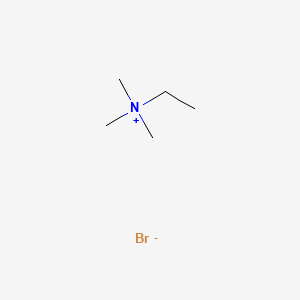 |
0.080 | ||
| ENC000713 | 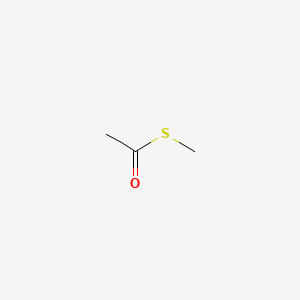 |
0.200 | D0X5SD | 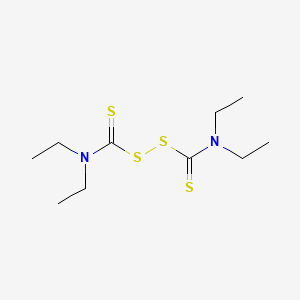 |
0.078 | ||
| ENC000522 | 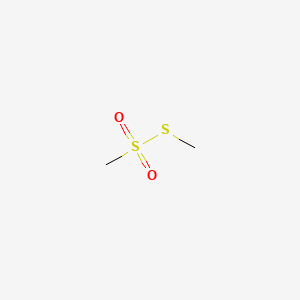 |
0.182 | D0F1GS | 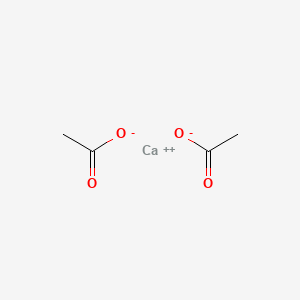 |
0.071 | ||
| ENC000656 | 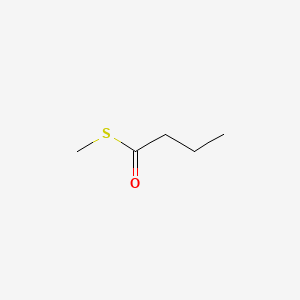 |
0.148 | D0E0SW | 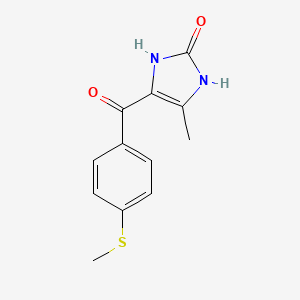 |
0.071 | ||
| ENC000689 | 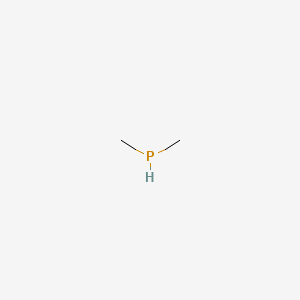 |
0.118 | D0C1QZ | 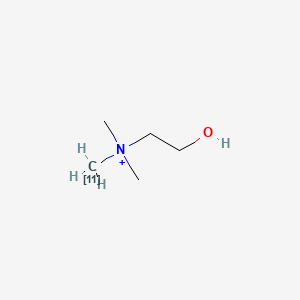 |
0.071 | ||
| ENC000286 | 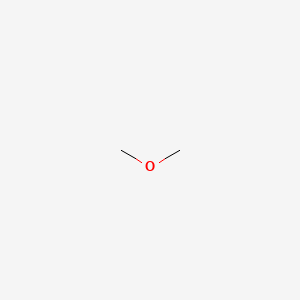 |
0.118 | D0Z4NI | 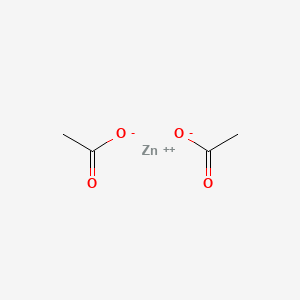 |
0.071 | ||
| ENC000355 | 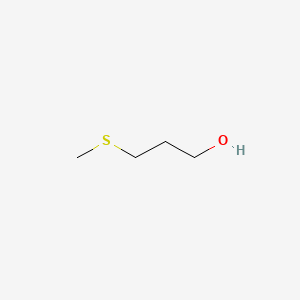 |
0.115 | D0XB8P | 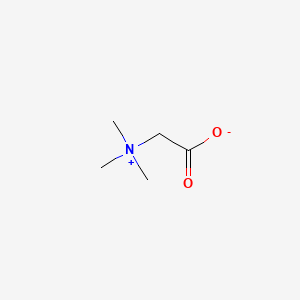 |
0.067 | ||
| ENC000524 | 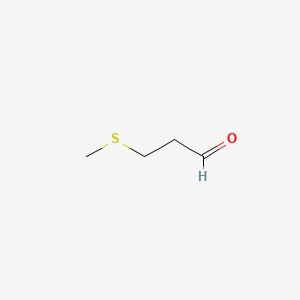 |
0.115 | D02KJX | 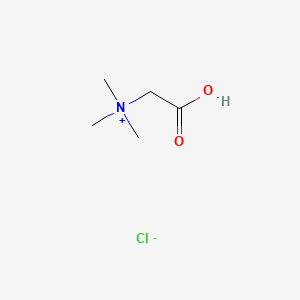 |
0.065 | ||
| ENC000132 | 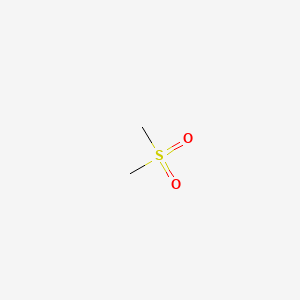 |
0.095 | D0ZK8H |  |
0.065 | ||