NPs Basic Information
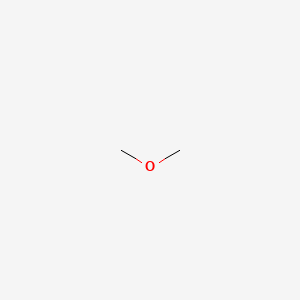
|
Name |
Dimethyl Ether
|
| Molecular Formula | C2H6O | |
| IUPAC Name* |
methoxymethane
|
|
| SMILES |
COC
|
|
| InChI |
InChI=1S/C2H6O/c1-3-2/h1-2H3
|
|
| InChIKey |
LCGLNKUTAGEVQW-UHFFFAOYSA-N
|
|
| Synonyms |
DIMETHYL ETHER; Methoxymethane; Methyl ether; Dimethyl oxide; Oxybismethane; Methane, oxybis-; 115-10-6; Methyl oxide; Wood ether; Dymel A; Ether, dimethyl; Ether, methyl; Demeon D; (CH3)2O; Methane, 1,1'-oxybis-; AM13FS69BX; CH3-O-CH3; 157621-61-9; CHEBI:28887; methylether; HSDB 354; Dimethyl ether, puriss., >=99.9% (GC); EINECS 204-065-8; UN1033; UNII-AM13FS69BX; monomethylether; Dimehtylether; Dymel; monomethyl ether; oxybis(methane); mono methyl ether; Methyloxoniomethanide; Dimethyl ether 99%; OMe2; (Methoxymethylidyne)radical; Dimethyl ether, >=99%; EC 204-065-8; DIMETHYL ETHER [MI]; 1,1'-OXYBISMETHANE; DIMETHYL ETHER [HSDB]; DIMETHYL ETHER [INCI]; Dimethyl ether, >=99.9%; DIMETHYL ETHER [VANDF]; CHEMBL119178; DIMETHYL ETHER [MART.]; DIMETHYL ETHER [WHO-DD]; DTXSID8026937; MFCD00008494; AKOS015915866; Dimethyl ether, puriss., >=99.8%; UN 1033; Dimethyl ether [UN1033] [Flammable gas]; FT-0628727; M0224; Q408050; 2F2
|
|
| CAS | 115-10-6 | |
| PubChem CID | 8254 | |
| ChEMBL ID | CHEMBL119178 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 46.07 | ALogp: | 0.1 |
| HBD: | 0 | HBA: | 1 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 9.2 | Aromatic Rings: | 0 |
| Heavy Atoms: | 3 | QED Weighted: | 0.389 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.136 | MDCK Permeability: | 0.00003930 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.026 |
| Human Intestinal Absorption (HIA): | 0.006 | 20% Bioavailability (F20%): | 0.08 |
| 30% Bioavailability (F30%): | 0.038 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.858 | Plasma Protein Binding (PPB): | 5.71% |
| Volume Distribution (VD): | 0.963 | Fu: | 88.69% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.214 | CYP1A2-substrate: | 0.947 |
| CYP2C19-inhibitor: | 0.042 | CYP2C19-substrate: | 0.866 |
| CYP2C9-inhibitor: | 0.006 | CYP2C9-substrate: | 0.257 |
| CYP2D6-inhibitor: | 0.006 | CYP2D6-substrate: | 0.646 |
| CYP3A4-inhibitor: | 0.006 | CYP3A4-substrate: | 0.293 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 7.436 | Half-life (T1/2): | 0.74 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.015 | Human Hepatotoxicity (H-HT): | 0.06 |
| Drug-inuced Liver Injury (DILI): | 0.036 | AMES Toxicity: | 0.067 |
| Rat Oral Acute Toxicity: | 0.585 | Maximum Recommended Daily Dose: | 0.199 |
| Skin Sensitization: | 0.4 | Carcinogencity: | 0.837 |
| Eye Corrosion: | 0.954 | Eye Irritation: | 0.991 |
| Respiratory Toxicity: | 0.074 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000382 | 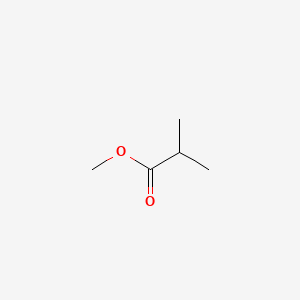 |
0.211 | D0A7MY |  |
0.185 | ||
| ENC000403 | 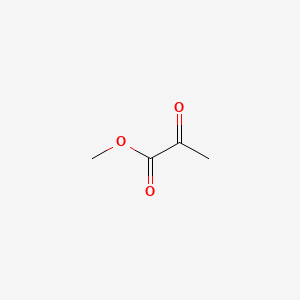 |
0.211 | D08HVE | 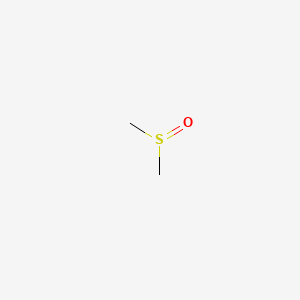 |
0.167 | ||
| ENC000689 | 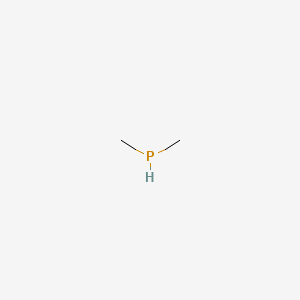 |
0.200 | D07SOO | 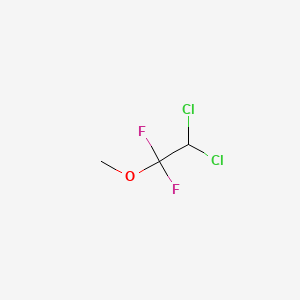 |
0.136 | ||
| ENC000719 | 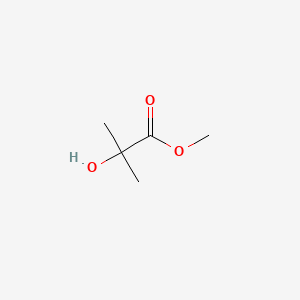 |
0.190 | D0ZK8H | 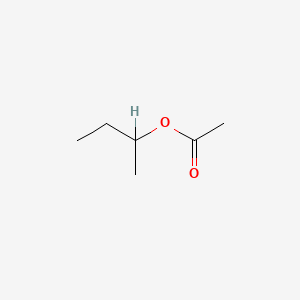 |
0.130 | ||
| ENC000234 | 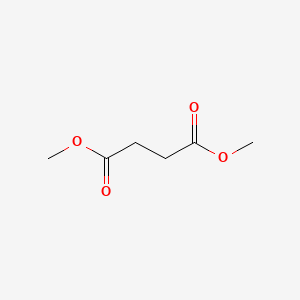 |
0.185 | D09GYT | 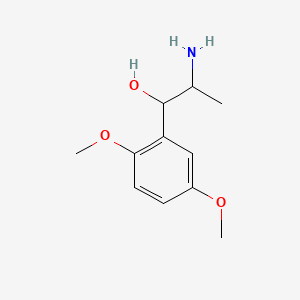 |
0.122 | ||
| ENC000312 | 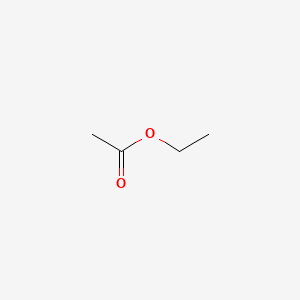 |
0.167 | D0FM2P |  |
0.120 | ||
| ENC000349 |  |
0.161 | D0U3IG |  |
0.111 | ||
| ENC000501 |  |
0.161 | D0Q9HF | 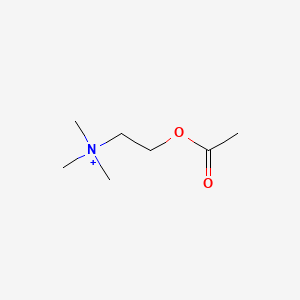 |
0.107 | ||
| ENC000736 | 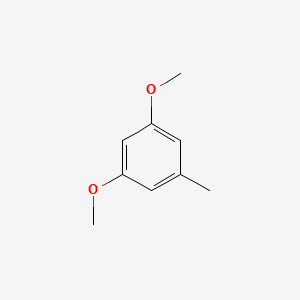 |
0.161 | D0U7BW | 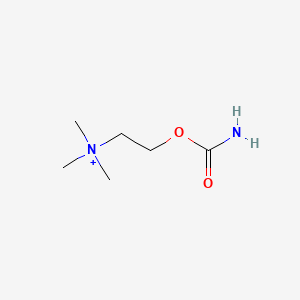 |
0.107 | ||
| ENC001351 | 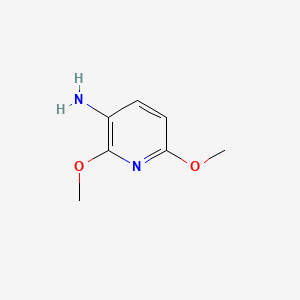 |
0.161 | D0OL6O | 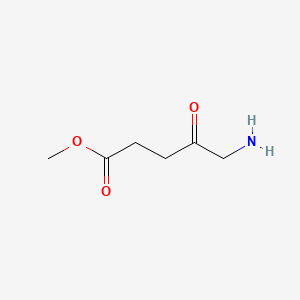 |
0.103 | ||