NPs Basic Information
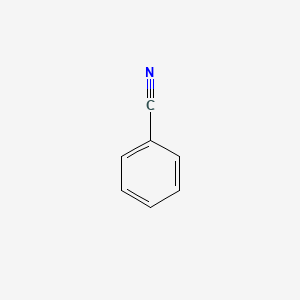
|
Name |
Benzonitrile
|
| Molecular Formula | C7H5N | |
| IUPAC Name* |
benzonitrile
|
|
| SMILES |
C1=CC=C(C=C1)C#N
|
|
| InChI |
InChI=1S/C7H5N/c8-6-7-4-2-1-3-5-7/h1-5H
|
|
| InChIKey |
JFDZBHWFFUWGJE-UHFFFAOYSA-N
|
|
| Synonyms |
BENZONITRILE; 100-47-0; Cyanobenzene; Phenyl cyanide; Benzenenitrile; Benzoic acid nitrile; Benzene, cyano-; Benzenecarbonitrile; Phenylcyanide; Fenylkyanid; Fenylkyanid [Czech]; NSC 8039; UN2224; AI3-24184; 9V9APP5H5S; C6H5-CN; CHEBI:27991; NSC-8039; Benzonitrile [UN2224] [Poison]; MFCD00001770; DSSTox_CID_1491; DSSTox_RID_76183; DSSTox_GSID_21491; benzonitril; CAS-100-47-0; HSDB 45; CCRIS 3184; EINECS 202-855-7; UNII-9V9APP5H5S; benzo nitrile; 4-cyanobenzene; benzonitrile solvent; WLN: NCR; BENZONITRILE [MI]; bmse000284; BENZONITRILE [HSDB]; EC 202-855-7; SCHEMBL6640; MLS002454387; CHEMBL15819; DTXSID7021491; NSC8039; Benzonitrile, anhydrous, >=99%; HMS3039F17; ZINC899417; Benzonitrile, for HPLC, 99.9%; Tox21_201982; Tox21_302979; Benzonitrile, ReagentPlus(R), 99%; STK398186; AKOS000120125; AM10697; UN 2224; NCGC00091747-01; NCGC00091747-02; NCGC00256387-01; NCGC00259531-01; LS-13256; SMR001372003; B0082; FT-0622719; EN300-19362; C09814; Q412567; J-000140; F1908-0163; Z104473628
|
|
| CAS | 100-47-0 | |
| PubChem CID | 7505 | |
| ChEMBL ID | CHEMBL15819 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 103.12 | ALogp: | 1.6 |
| HBD: | 0 | HBA: | 1 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 23.8 | Aromatic Rings: | 1 |
| Heavy Atoms: | 8 | QED Weighted: | 0.493 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.179 | MDCK Permeability: | 0.00003120 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.004 | 20% Bioavailability (F20%): | 0.06 |
| 30% Bioavailability (F30%): | 0.004 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.806 | Plasma Protein Binding (PPB): | 71.83% |
| Volume Distribution (VD): | 2.338 | Fu: | 17.84% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.956 | CYP1A2-substrate: | 0.502 |
| CYP2C19-inhibitor: | 0.375 | CYP2C19-substrate: | 0.428 |
| CYP2C9-inhibitor: | 0.14 | CYP2C9-substrate: | 0.477 |
| CYP2D6-inhibitor: | 0.011 | CYP2D6-substrate: | 0.403 |
| CYP3A4-inhibitor: | 0.015 | CYP3A4-substrate: | 0.316 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 8.75 | Half-life (T1/2): | 0.816 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.097 | Human Hepatotoxicity (H-HT): | 0.898 |
| Drug-inuced Liver Injury (DILI): | 0.276 | AMES Toxicity: | 0.09 |
| Rat Oral Acute Toxicity: | 0.087 | Maximum Recommended Daily Dose: | 0.06 |
| Skin Sensitization: | 0.38 | Carcinogencity: | 0.605 |
| Eye Corrosion: | 0.968 | Eye Irritation: | 0.995 |
| Respiratory Toxicity: | 0.291 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000052 | 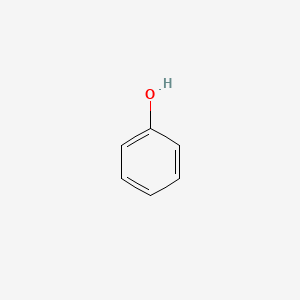 |
0.483 | D05OIS | 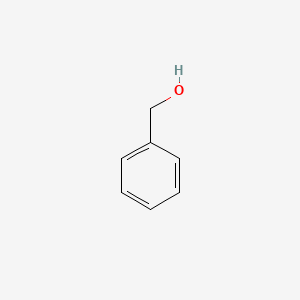 |
0.438 | ||
| ENC000064 | 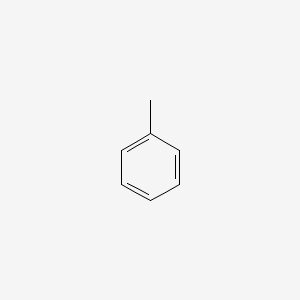 |
0.483 | D0X9RY | 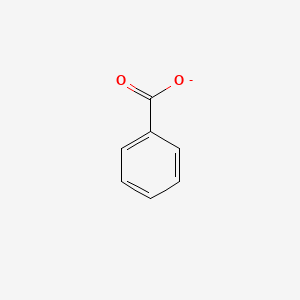 |
0.412 | ||
| ENC000014 | 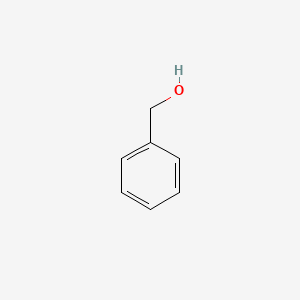 |
0.438 | D0H0HJ | 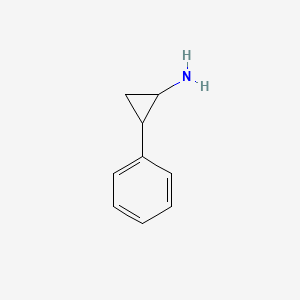 |
0.378 | ||
| ENC000203 |  |
0.438 | D0T3LF | 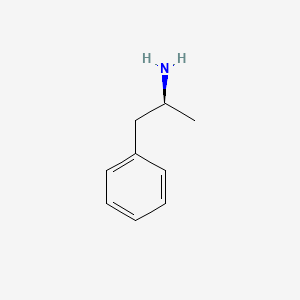 |
0.378 | ||
| ENC000207 | 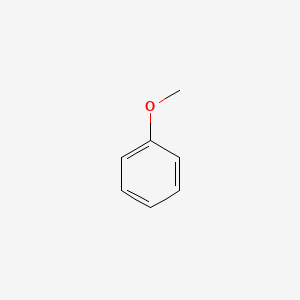 |
0.438 | D05BMG | 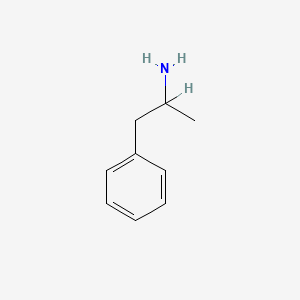 |
0.378 | ||
| ENC000204 | 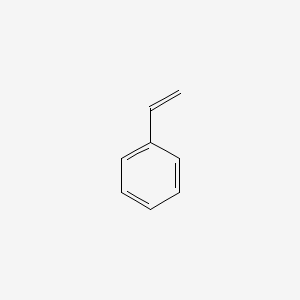 |
0.438 | D0P9AC | 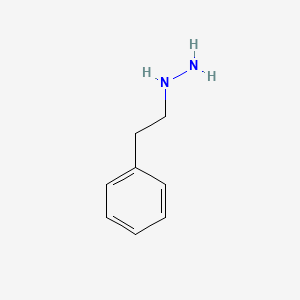 |
0.368 | ||
| ENC000012 | 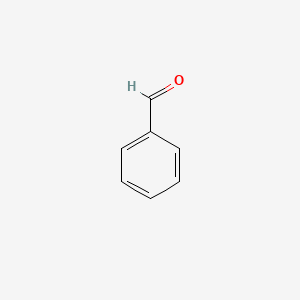 |
0.438 | D0U0RZ | 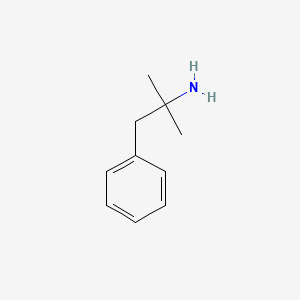 |
0.359 | ||
| ENC000205 | 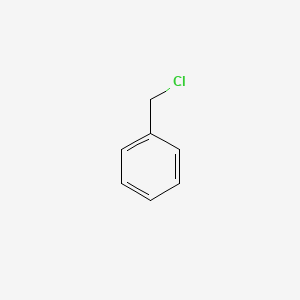 |
0.438 | D0R0UJ | 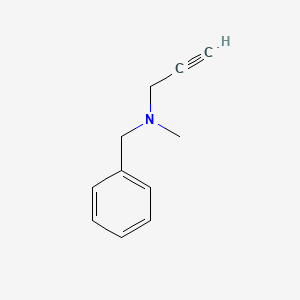 |
0.357 | ||
| ENC000076 | 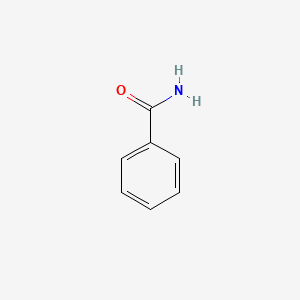 |
0.412 | D0P6UB | 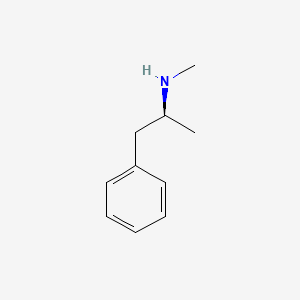 |
0.350 | ||
| ENC000013 | 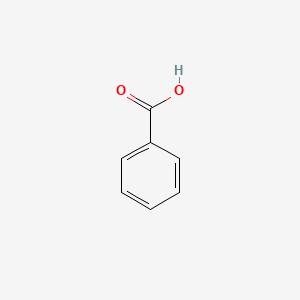 |
0.412 | D01ZJK | 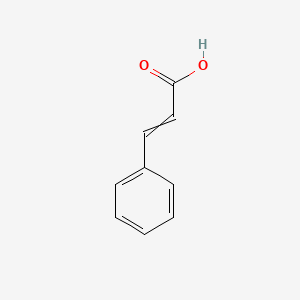 |
0.350 | ||