NPs Basic Information
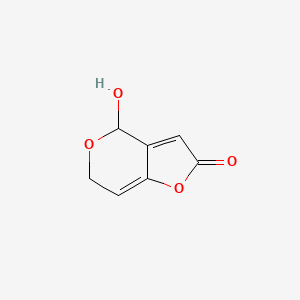
|
Name |
Patulin
|
| Molecular Formula | C7H6O4 | |
| IUPAC Name* |
4-hydroxy-4,6-dihydrofuro[3,2-c]pyran-2-one
|
|
| SMILES |
C1C=C2C(=CC(=O)O2)C(O1)O
|
|
| InChI |
InChI=1S/C7H6O4/c8-6-3-4-5(11-6)1-2-10-7(4)9/h1,3,7,9H,2H2
|
|
| InChIKey |
ZRWPUFFVAOMMNM-UHFFFAOYSA-N
|
|
| Synonyms |
patulin; 149-29-1; Clavacin; Clavatin; Expansin; Patuline; Expansine; Claviformin; Claviform; Clairformin; Leucopin; Terinin; Gigantin; Mycoin; Mycoin C3; Penicidin; Tercinin; Mycoine C3; Mycosin; Penatin; Mycoin C; 4-Hydroxy-4H-furo[3,2-c]pyran-2(6H)-one; 4-hydroxy-4,6-dihydrofuro[3,2-c]pyran-2-one; 4H-Furo[3,2-c]pyran-2(6H)-one, 4-hydroxy-; Sch-351633; NSC 8120; NSC 32951; 4-Hydroxy-4H-furo(3,2-C)pyran-2(6H)-one; 4H-Furo(3,2-c)pyran-2(6H)-one, 4-hydroxy-; (2,4-Dihydroxy-2H-pyran-3(6H)-ylidene)acetic acid, 3,4-lactone; 4,6-Dihydro-4-hydroxy-2H-furo(3,2-c)pyran-2-one; NSC8120; NSC32951; SCH 351633; (2,4-Dihydroxy-2H-pyran-3(6H)-ylidene)acetic acid-3,4-lactone; Acetic acid, (2,4-dihydroxy-2H-pyran-3(6H)-ylidene)-, 3,4-lactone; 4,6-dihydro-4-hydroxy-2H-furo[3,2-c]pyran-2-one; 95X2BV4W8R; CHEMBL294018; CHEBI:74926; NSC-8120; NSC-32951; (2, 3,4-lactone; 4-Hydroxy-4,6-dihydrofuro[4,5-c]pyran-2-one; 4H-Furo[3, 4-hydroxy-; 4-hydroxy-4,6-dihydro-2H-furo[3,2-c]pyran-2-one; 2,.alpha.-acetic acid, 3,4-lactone; Patulin 100 microg/mL in Acetonitrile; WLN: T56 BOV GO IU & TJ FQ; CCRIS 4940; HSDB 3522; SR-05000002238; EINECS 205-735-2; MFCD00005858; BRN 0149675; UNII-95X2BV4W8R; Clavicin; Clavitin; Expansion; 2H-Pyran-.delta.(sup 3(6H), 2,4-dihydroxy-, 3,4-lactone; Acetic acid,4-dihydroxy-2H-pyran-3(6H)-ylidene)-, 3,4-lactone; Gigantic acid?; 4H-Furo(3,3-c)pyran-2(6H)-one, 4-hydroxy-; DL-PATULIN; Patulin-[13C3]; Spectrum_000015; starbld0009637; Antibiotic YS 1649; PATULIN [HSDB]; PATULIN [IARC]; PATULIN [MI]; 2,4-Dihydroxy-2H-pyran-delta-3(6H),alpha-acetic acid-3,4-lactone; Spectrum3_000796; Spectrum4_000753; Spectrum5_001659; PATULIN 13C3; 2H-Pyran-delta(sup 3(6H),alpha)-acetic acid, 2,4-dihydroxy-, 3,4-lactone; Antibiotic Sch 351633; Patulin, reference material; Neuro_000008; Clairformin;Patuline;Clavatin; SCHEMBL29056; BSPBio_002532; KBioGR_001106; KBioSS_000355; 5-18-03-00005 (Beilstein Handbook Reference); DivK1c_000438; SPECTRUM1503904; MEGxm0_000442; DTXSID2021101; ACon1_002106; HMS501F20; KBio1_000438; KBio2_000355; KBio2_002923; KBio2_005491; KBio3_001752; NINDS_000438; HMS1923M19; Patulin, >=98.0% (HPLC); BCP29227; EX-A5478; HY-N6779; BDBM50158841; AKOS015904103; BS-1260; CCG-208451; DB15586; YS 1649; IDI1_000438; SMP1_000230; NCGC00095272-01; NCGC00095272-02; NCGC00095272-03; NCGC00095272-04; NCGC00095272-05; NCI60_041782; DB-042987; 4-Hydroxy-4H,6H-furo[3,2-c]pyran-2-one; CS-0083018; FT-0631849; FT-0673527; 4-Hydroxy-4H-Furo(3,3-c)pyran-2(6H)-one; 4-hydroxy-2H,4H,6H-furo[3,2-c]pyran-2-one; Q414526; J-008576; SR-05000002238-2; SR-05000002238-3; 2,4-Dihydroxy-2H-pyran-alpha -acetic acid, 3, 4-lactone; 4-Hydroxy-4H-furo[3,2-c]pyran-2(6H)-one, 9CI, 8CI; (+/-)-4-HYDROXY-4H-FURO(3,2-C)PYRAN-2(6H)-ONE; 2H-Pyran-.delta.(sup 3(6H), 2,4-dihydroxy-,3,4-lactone; 2H-Pyran-alpha )-acetic acid, 2, 4-dihydroxy-,3,4-lactone; 2H-Pyran-alpha )-acetic acid, 2,4-dihydroxy-, 3,4-lactone; 2,4-Dihydroxy-2H-pyran-.delta.-3(6H),.alpha.-acetic acid, 3,4-lactone; 2H-Pyran-.delta.(3(6H),.alpha.)-acetic acid, 2,4-dihydroxy-,3,4-lactone; 247172-18-5
|
|
| CAS | 149-29-1 | |
| PubChem CID | 4696 | |
| ChEMBL ID | CHEMBL294018 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 154.12 | ALogp: | -1.0 |
| HBD: | 1 | HBA: | 4 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 55.8 | Aromatic Rings: | 2 |
| Heavy Atoms: | 11 | QED Weighted: | 0.504 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.683 | MDCK Permeability: | 0.00003250 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.016 | 20% Bioavailability (F20%): | 0.01 |
| 30% Bioavailability (F30%): | 0.705 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.235 | Plasma Protein Binding (PPB): | 80.23% |
| Volume Distribution (VD): | 1.274 | Fu: | 29.03% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.547 | CYP1A2-substrate: | 0.448 |
| CYP2C19-inhibitor: | 0.075 | CYP2C19-substrate: | 0.066 |
| CYP2C9-inhibitor: | 0.056 | CYP2C9-substrate: | 0.764 |
| CYP2D6-inhibitor: | 0.025 | CYP2D6-substrate: | 0.483 |
| CYP3A4-inhibitor: | 0.013 | CYP3A4-substrate: | 0.163 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 14.074 | Half-life (T1/2): | 0.875 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.011 | Human Hepatotoxicity (H-HT): | 0.344 |
| Drug-inuced Liver Injury (DILI): | 0.864 | AMES Toxicity: | 0.38 |
| Rat Oral Acute Toxicity: | 0.052 | Maximum Recommended Daily Dose: | 0.021 |
| Skin Sensitization: | 0.237 | Carcinogencity: | 0.53 |
| Eye Corrosion: | 0.022 | Eye Irritation: | 0.653 |
| Respiratory Toxicity: | 0.514 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC006134 | 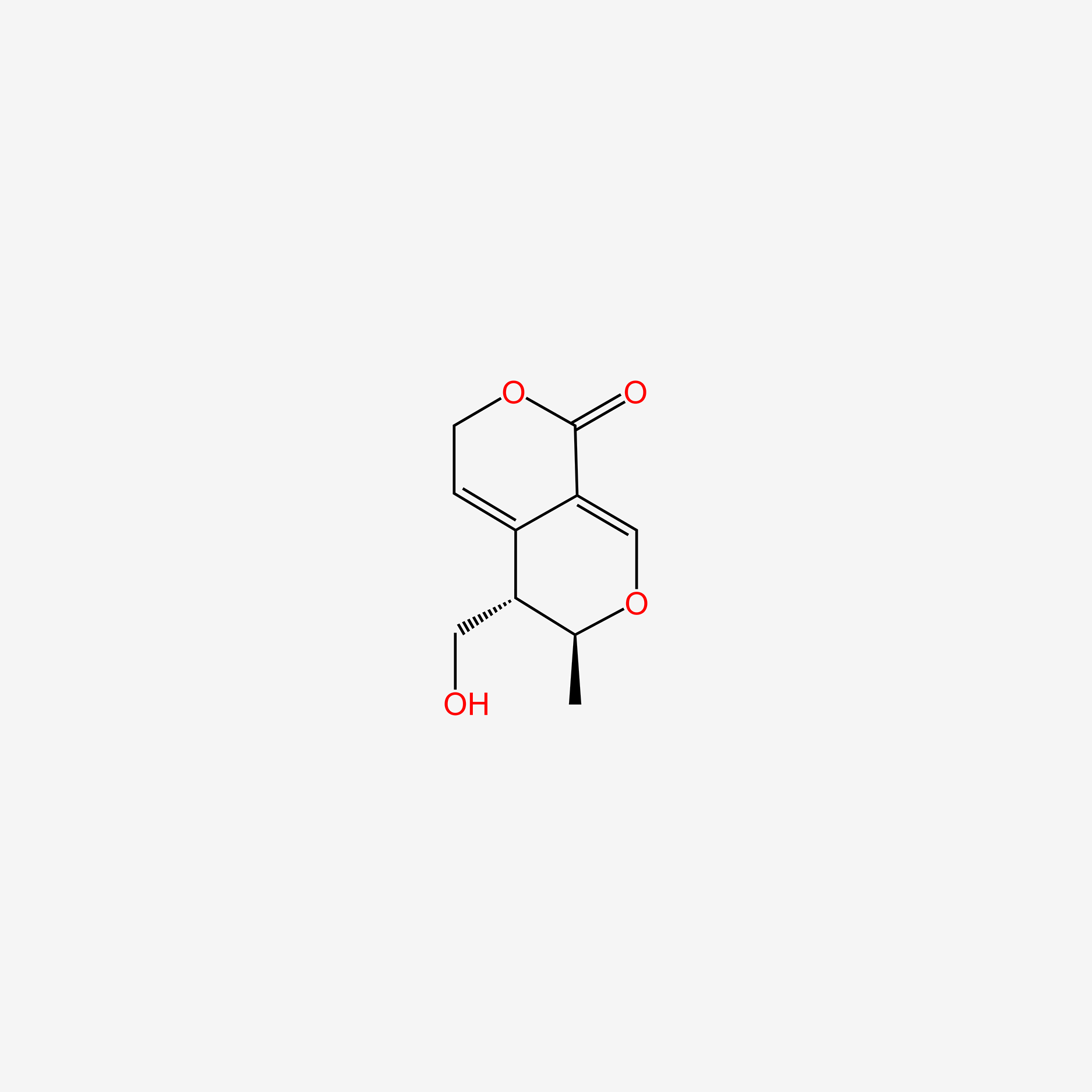 |
0.273 | D0L1WV | 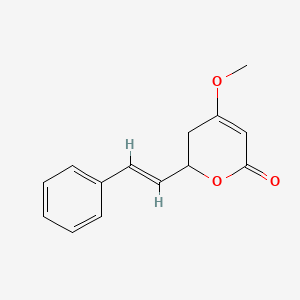 |
0.176 | ||
| ENC006135 | 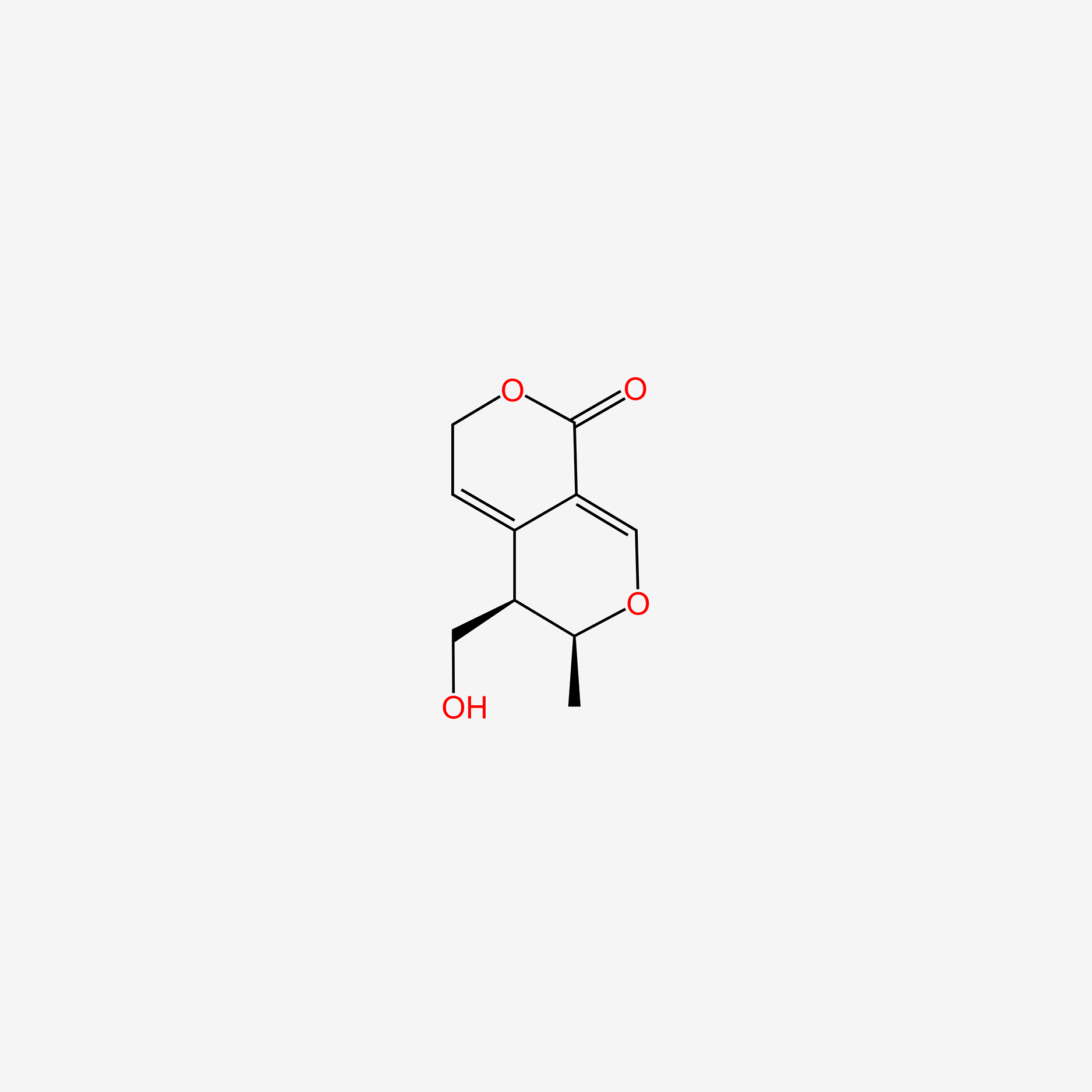 |
0.273 | D09PZO | 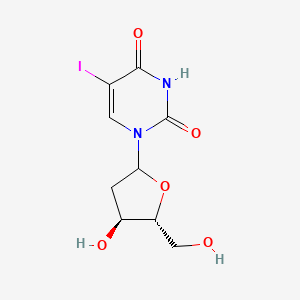 |
0.167 | ||
| ENC006136 | 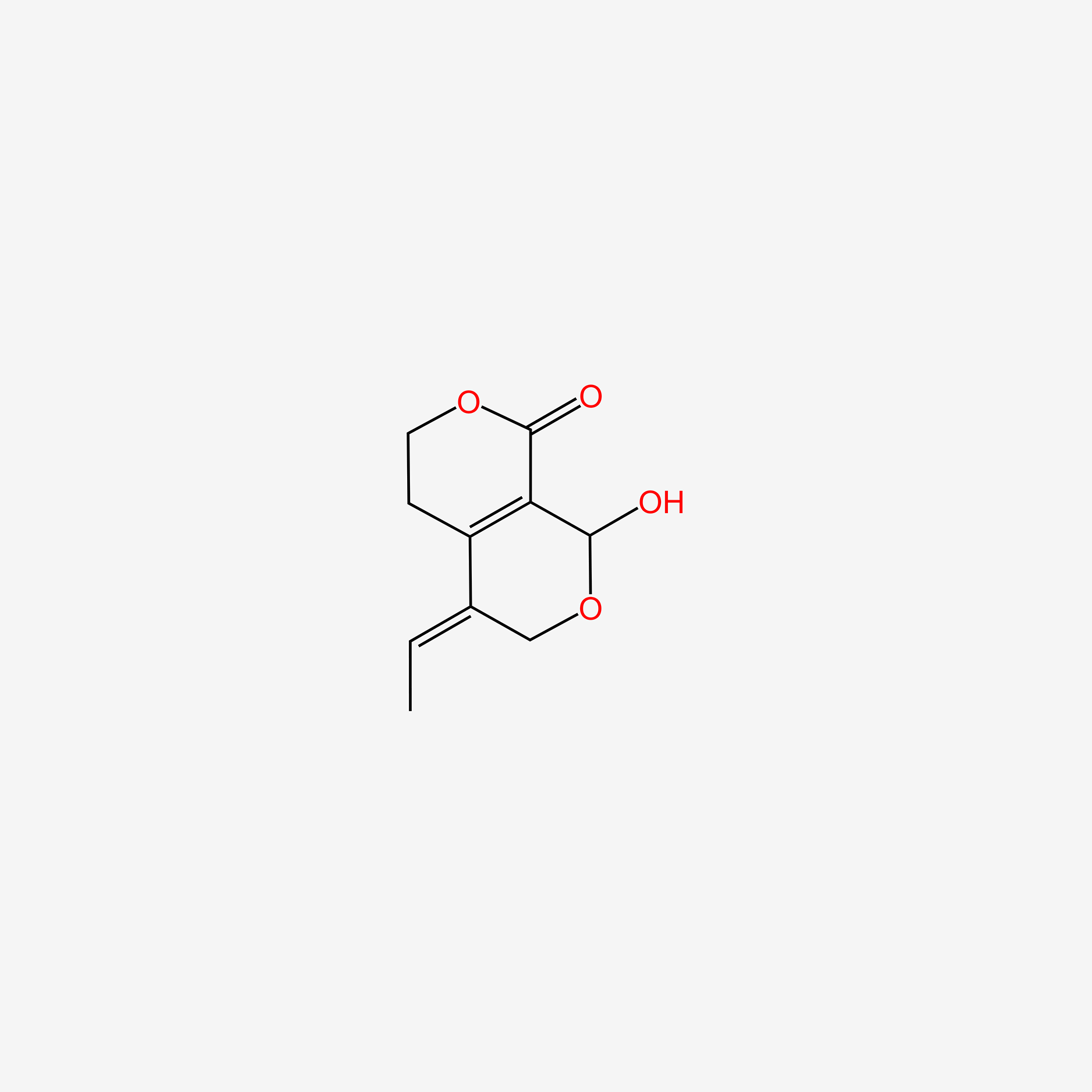 |
0.228 | D0CL9S | 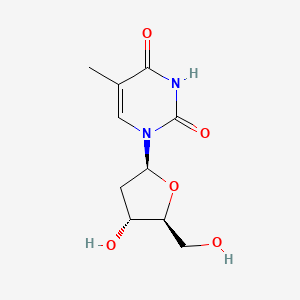 |
0.167 | ||
| ENC005475 |  |
0.226 | D0TS1Z | 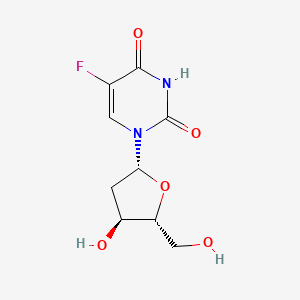 |
0.167 | ||
| ENC004626 | 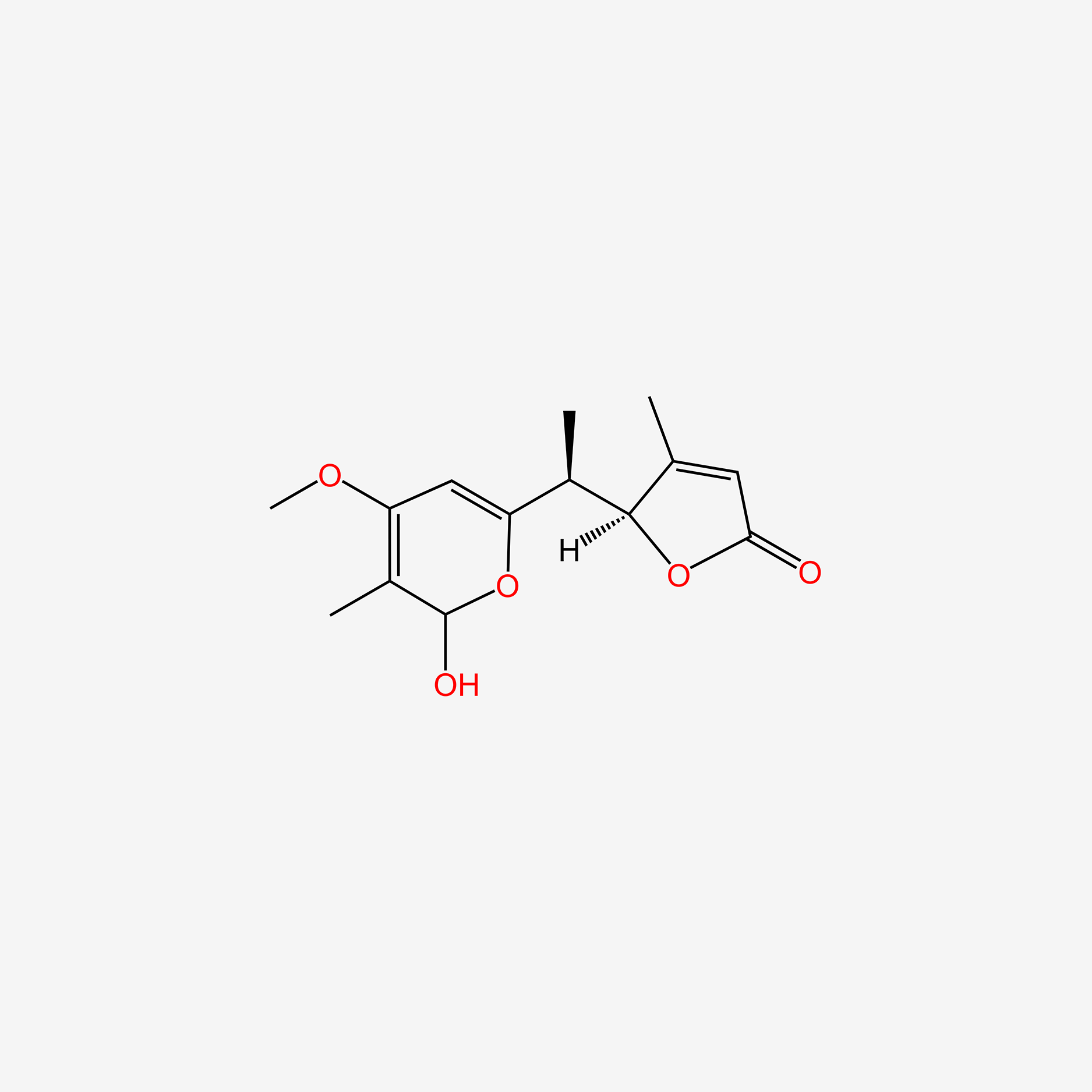 |
0.224 | D0N0OU | 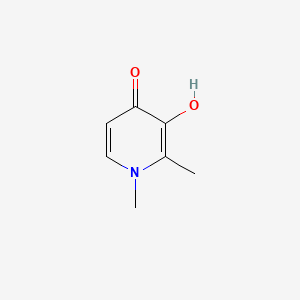 |
0.163 | ||
| ENC004166 | 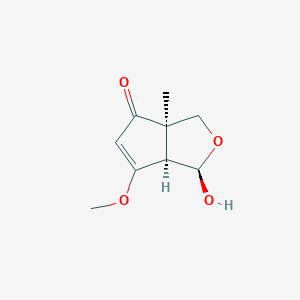 |
0.222 | D06FDR | 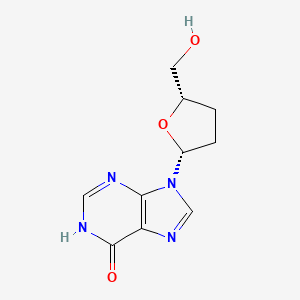 |
0.159 | ||
| ENC004165 | 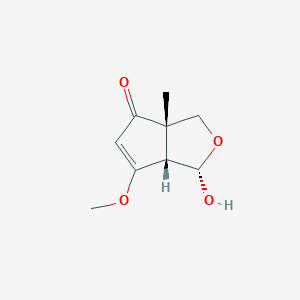 |
0.222 | D0Z9QR | 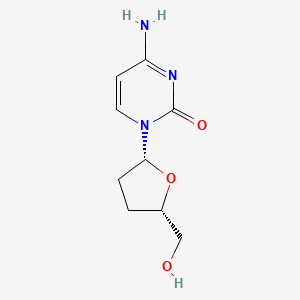 |
0.159 | ||
| ENC002485 | 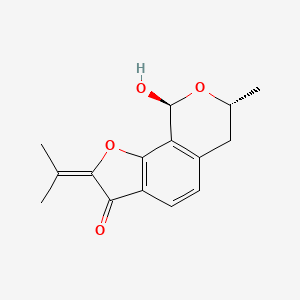 |
0.221 | D07TQV | 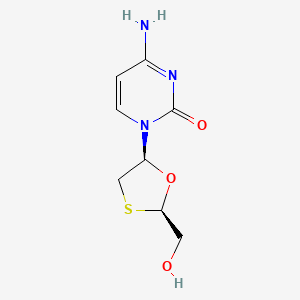 |
0.159 | ||
| ENC002838 | 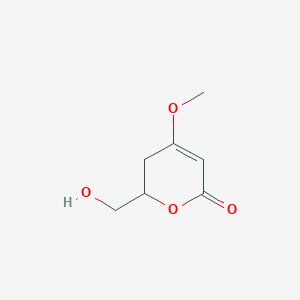 |
0.220 | D07GRH | 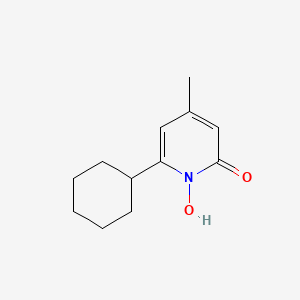 |
0.159 | ||
| ENC005200 | 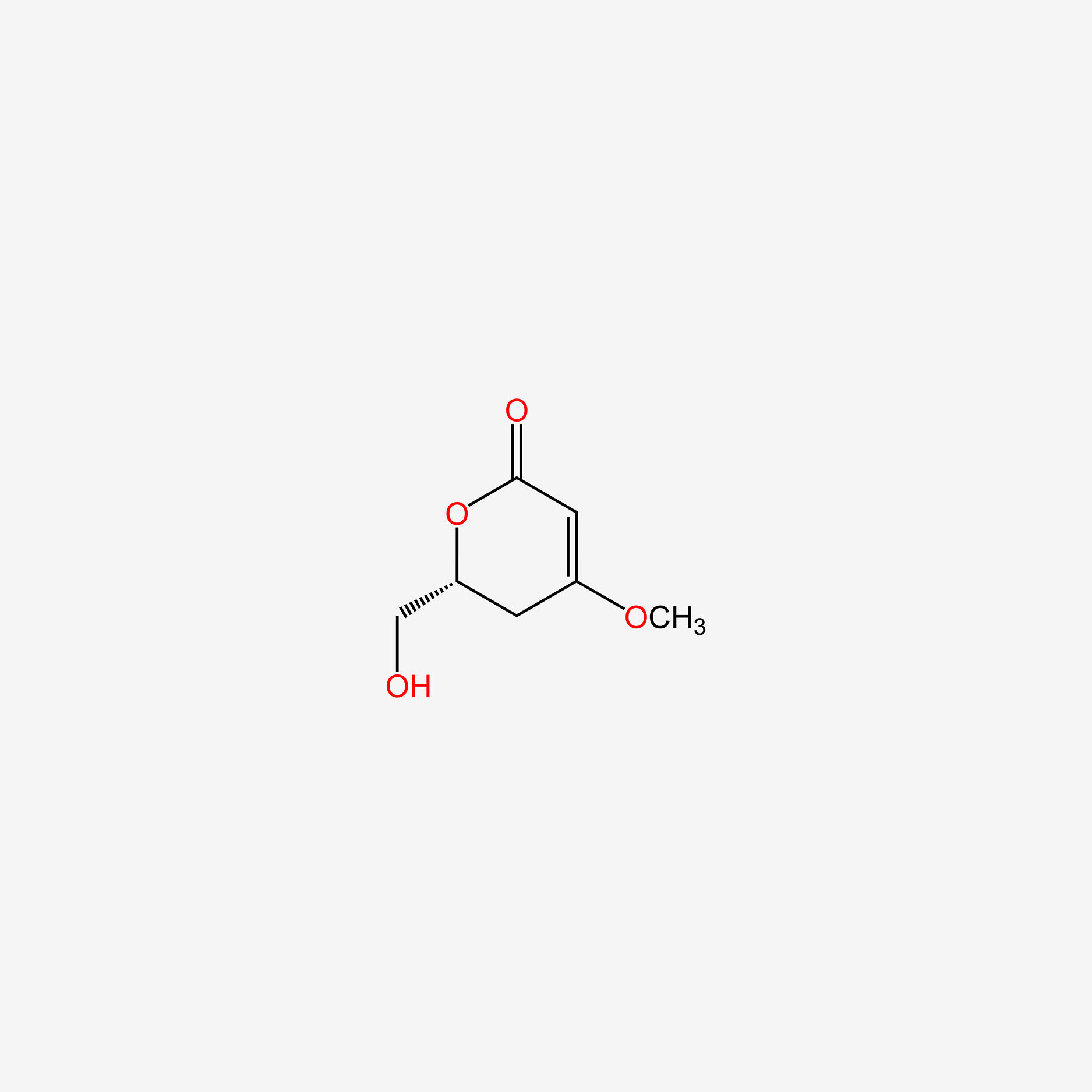 |
0.220 | D0A2AJ | 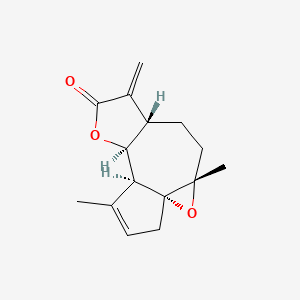 |
0.157 | ||