NPs Basic Information
|
Name |
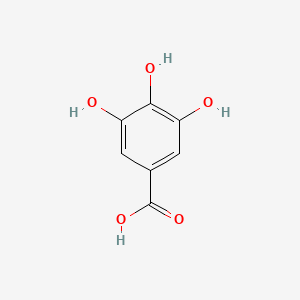
Gallic Acid
|
| Molecular Formula | C7H6O5 | |
| IUPAC Name* |
3,4,5-trihydroxybenzoic acid
|
|
| SMILES |
C1=C(C=C(C(=C1O)O)O)C(=O)O
|
|
| InChI |
InChI=1S/C7H6O5/c8-4-1-3(7(11)12)2-5(9)6(4)10/h1-2,8-10H,(H,11,12)
|
|
| InChIKey |
LNTHITQWFMADLM-UHFFFAOYSA-N
|
|
| Synonyms |
Gallic acid; 3,4,5-Trihydroxybenzoic acid; 149-91-7; gallate; Benzoic acid, 3,4,5-trihydroxy-; Gallic acid, tech.; GALOP; Pyrogallol-5-carboxylic acid; Kyselina gallova; HSDB 2117; 3,4,5-Trihydroxybenzoate; MFCD00002510; CCRIS 5523; NSC 674319; CHEBI:30778; AI3-16412; Kyselina 3,4,5-trihydroxybenzoova; NSC-20103; NSC-674319; CHEMBL288114; 3,4,5-trihydroxy-Benzoic acid; 632XD903SP; NSC20103; NSC674319; NCGC00091125-01; DSSTox_CID_650; DSSTox_RID_75711; DSSTox_GSID_20650; GALLIC ACID ANHYDROUS; Kyselina gallova [Czech]; 31387-49-2; CAS-149-91-7; Gallic acid [NF]; SR-05000001537; EINECS 205-749-9; NSC 20103; BRN 2050274; gallic-acid; UNII-632XD903SP; Kyselina 3,4,5-trihydroxybenzoova [Czech]; Gallic acid tech.; Gallic Acid, F; GDE; (?)-Gallic acid; Spectrum_000342; 3,4,5-Trihydroxybenzoic acid, anhydrous; SpecPlus_000307; 5-Trihydroxybenzoic acid; Spectrum2_000399; Spectrum3_000254; Spectrum4_001544; Spectrum5_000108; GALLIC ACID [MI]; bmse000389; 3,5-Trihydroxybenzoic acid; GALLIC ACID [HSDB]; GALLIC ACID [INCI]; WLN: QVR CQ DQ EQ; 3,4,5-trihydroxy-Benzoate; Oprea1_087792; SCHEMBL15012; 3,4,5-Trihydroxybenzoicacid; BSPBio_001668; KBioGR_002008; KBioSS_000822; SPECTRUM210369; 3-10-00-02070 (Beilstein Handbook Reference); BIDD:ER0374; DivK1c_006403; GALLIC ACID [WHO-DD]; SPBio_000617; 3,4,5-Trihydroxybenzoate, X; Benzoic acid,4,5-trihydroxy-; GTPL5549; ZINC1504; 3,4,5-Trihydroxybenzoic acid;; DTXSID0020650; KBio1_001347; KBio2_000822; KBio2_003390; KBio2_005958; KBio3_001168; CPD-183; Gallic acid, puriss., 98.0%; HMS1923K07; HMS2091A07; Pharmakon1600-00210369; BCP18127; HY-N0523; NSC36997; Tox21_111089; Tox21_202515; BBL009937; BDBM50085536; CCG-38670; NSC-36997; NSC755825; s4603; STK298718; AKOS000119625; Tox21_111089_1; AC-1206; CS-8191; NSC-755825; PS-8710; SDCCGMLS-0066503.P001; NCGC00091125-02; NCGC00091125-03; NCGC00091125-04; NCGC00091125-05; NCGC00091125-07; NCGC00260064-01; DA-33612; SY038078; SBI-0052184.P002; Gallic acid, 97.5-102.5% (titration); FT-0626592; G0011; EN300-21542; C01424; D85056; 3,4,5-trihydroxybenzoic acid (ACD/Name 4.0); AB00052697_03; Q375837; Q-201146; SR-05000001537-1; SR-05000001537-2; SR-05000001537-3; BRD-K77345217-001-01-9; F1908-0156; Gallic acid, certified reference material, TraceCERT(R); Z104501122; 78563C7D-0E2D-4766-A8EA-670A03C78FCF; 137657-43-3
|
|
| CAS | 149-91-7 | |
| PubChem CID | 370 | |
| ChEMBL ID | CHEMBL288114 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 170.12 | ALogp: | 0.7 |
| HBD: | 4 | HBA: | 5 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 98.0 | Aromatic Rings: | 1 |
| Heavy Atoms: | 12 | QED Weighted: | 0.469 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.728 | MDCK Permeability: | 0.00000511 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.003 |
| Human Intestinal Absorption (HIA): | 0.085 | 20% Bioavailability (F20%): | 0.964 |
| 30% Bioavailability (F30%): | 0.995 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.099 | Plasma Protein Binding (PPB): | 53.49% |
| Volume Distribution (VD): | 0.466 | Fu: | 33.59% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.023 | CYP1A2-substrate: | 0.075 |
| CYP2C19-inhibitor: | 0.026 | CYP2C19-substrate: | 0.039 |
| CYP2C9-inhibitor: | 0.188 | CYP2C9-substrate: | 0.061 |
| CYP2D6-inhibitor: | 0.008 | CYP2D6-substrate: | 0.115 |
| CYP3A4-inhibitor: | 0.034 | CYP3A4-substrate: | 0.019 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 10.108 | Half-life (T1/2): | 0.947 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.017 | Human Hepatotoxicity (H-HT): | 0.433 |
| Drug-inuced Liver Injury (DILI): | 0.852 | AMES Toxicity: | 0.053 |
| Rat Oral Acute Toxicity: | 0.03 | Maximum Recommended Daily Dose: | 0.01 |
| Skin Sensitization: | 0.871 | Carcinogencity: | 0.024 |
| Eye Corrosion: | 0.488 | Eye Irritation: | 0.908 |
| Respiratory Toxicity: | 0.381 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000193 | 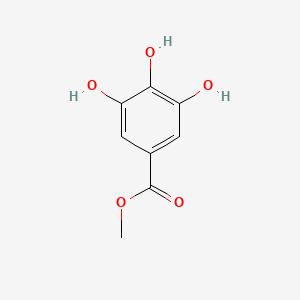 |
0.667 | D0V9EN | 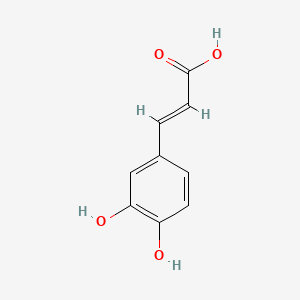 |
0.347 | ||
| ENC000002 | 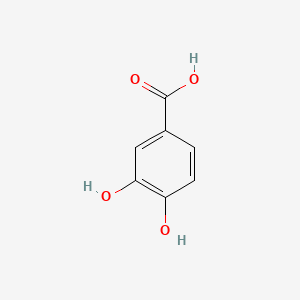 |
0.538 | D0I3RO | 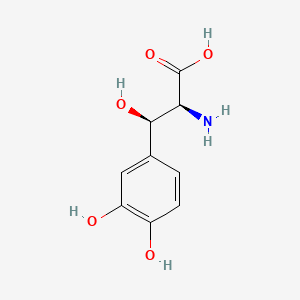 |
0.346 | ||
| ENC000674 | 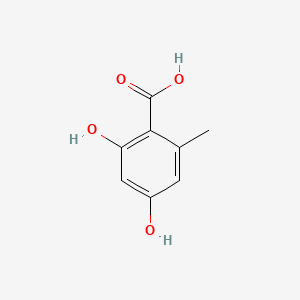 |
0.442 | D0C4YC | 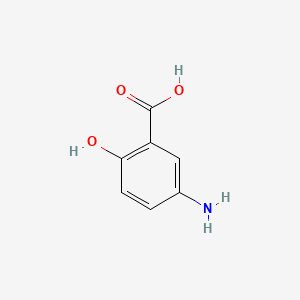 |
0.333 | ||
| ENC002875 | 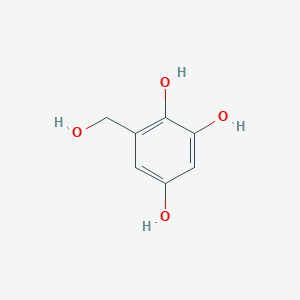 |
0.429 | D08HVR | 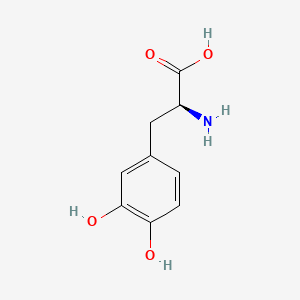 |
0.333 | ||
| ENC000367 | 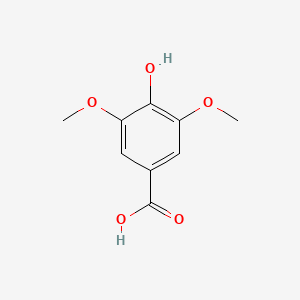 |
0.417 | D01WJL | 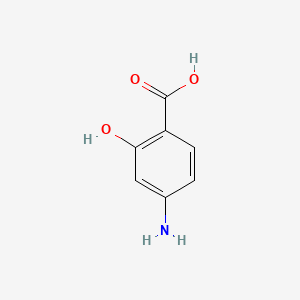 |
0.333 | ||
| ENC004830 | 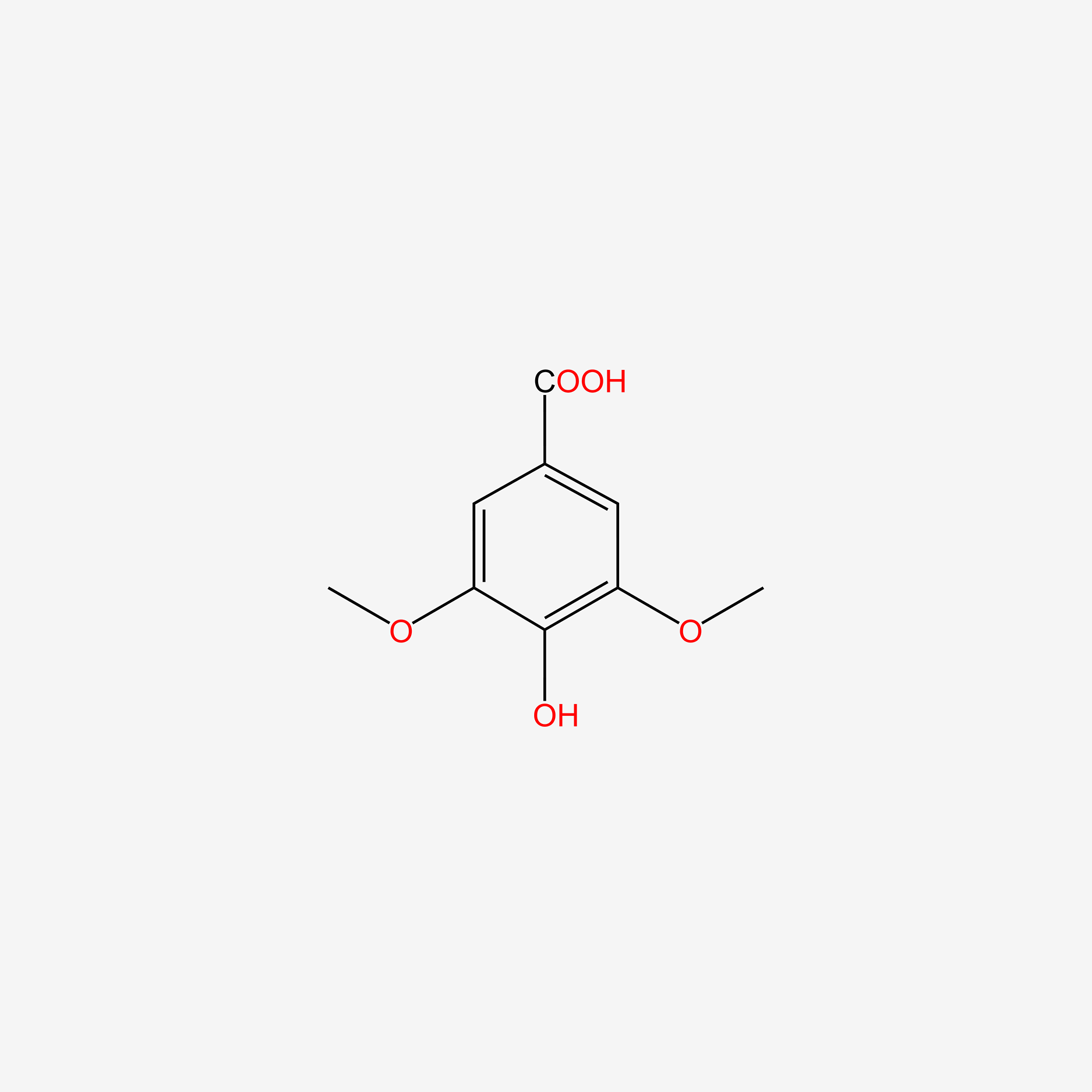 |
0.417 | D0BA6T | 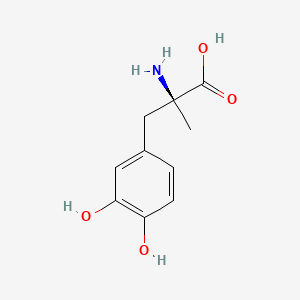 |
0.321 | ||
| ENC000097 | 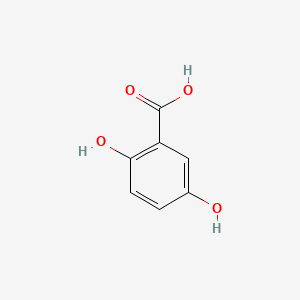 |
0.395 | D07EXH | 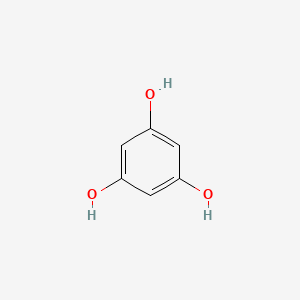 |
0.310 | ||
| ENC000069 | 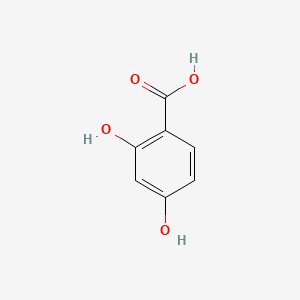 |
0.395 | D0Y7PG | 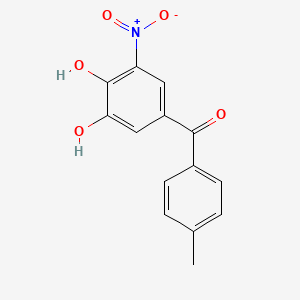 |
0.308 | ||
| ENC001445 | 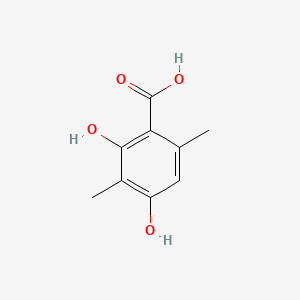 |
0.391 | D0P7JZ | 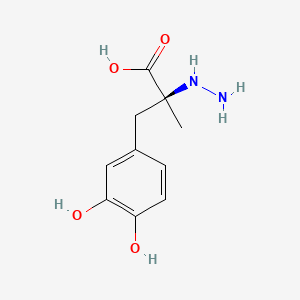 |
0.304 | ||
| ENC000060 | 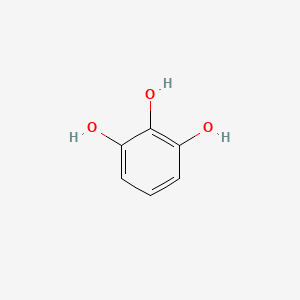 |
0.375 | D00KRE | 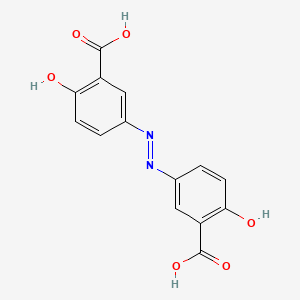 |
0.300 | ||