NPs Basic Information
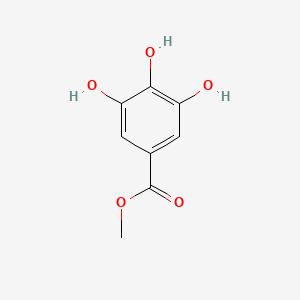
|
Name |
Methyl gallate
|
| Molecular Formula | C8H8O5 | |
| IUPAC Name* |
methyl 3,4,5-trihydroxybenzoate
|
|
| SMILES |
COC(=O)C1=CC(=C(C(=C1)O)O)O
|
|
| InChI |
InChI=1S/C8H8O5/c1-13-8(12)4-2-5(9)7(11)6(10)3-4/h2-3,9-11H,1H3
|
|
| InChIKey |
FBSFWRHWHYMIOG-UHFFFAOYSA-N
|
|
| Synonyms |
METHYL GALLATE; Methyl 3,4,5-trihydroxybenzoate; 99-24-1; Gallic acid methyl ester; Methylgallate; Benzoic acid, 3,4,5-trihydroxy-, methyl ester; Gallic acid, methyl ester; methyl galloate; NSC 363001; GALLINCIN; 3,4,5-Trihydroxybenzoic acid methyl ester; AI3-00861; 3,4,5-Trihydroxy-benzoic acid methyl ester; MFCD00002194; CHEMBL65675; 623D3XG80C; NSC363001; NSC-363001; CCRIS 5567; EINECS 202-741-7; BRN 2113180; UNII-623D3XG80C; Methyl gallate;; methyl-3,4,5-trihydroxybenzoate; Gallic acid methyl; cid_7428; 6-methylthiomorpholin-3-one; SCHEMBL39513; 4-10-00-01998 (Beilstein Handbook Reference); MLS000574912; DTXSID3059189; ZINC21789; CHEBI:145828; HMS2210A23; HMS3346G16; ACT10740; HY-N2010; BBL010505; BDBM50187133; s3790; STL146151; methyl 3,4,5-tris(oxidanyl)benzoate; AKOS000277450; GALLIC ACID METHYL ESTER [MI]; CCG-266463; Methyl 3,4,5-trihydroxybenzoate, 98%; NCGC00247606-01; NCGC00247606-02; AC-11364; AC-34481; AS-14808; SMR000156263; SY011144; DB-057768; CS-0018330; FT-0626596; G0017; EN300-393116; A845986; CS-008/03903045; W-100047; Q15425833; Z1255434713; M G
|
|
| CAS | 99-24-1 | |
| PubChem CID | 7428 | |
| ChEMBL ID | CHEMBL65675 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 184.15 | ALogp: | 0.9 |
| HBD: | 3 | HBA: | 5 |
| Rotatable Bonds: | 2 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 87.0 | Aromatic Rings: | 1 |
| Heavy Atoms: | 13 | QED Weighted: | 0.447 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.036 | MDCK Permeability: | 0.00001130 |
| Pgp-inhibitor: | 0.002 | Pgp-substrate: | 0.002 |
| Human Intestinal Absorption (HIA): | 0.025 | 20% Bioavailability (F20%): | 0.323 |
| 30% Bioavailability (F30%): | 0.955 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.154 | Plasma Protein Binding (PPB): | 85.38% |
| Volume Distribution (VD): | 0.494 | Fu: | 12.38% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.602 | CYP1A2-substrate: | 0.648 |
| CYP2C19-inhibitor: | 0.045 | CYP2C19-substrate: | 0.057 |
| CYP2C9-inhibitor: | 0.233 | CYP2C9-substrate: | 0.437 |
| CYP2D6-inhibitor: | 0.07 | CYP2D6-substrate: | 0.222 |
| CYP3A4-inhibitor: | 0.109 | CYP3A4-substrate: | 0.107 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 16.935 | Half-life (T1/2): | 0.942 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.021 | Human Hepatotoxicity (H-HT): | 0.045 |
| Drug-inuced Liver Injury (DILI): | 0.674 | AMES Toxicity: | 0.225 |
| Rat Oral Acute Toxicity: | 0.012 | Maximum Recommended Daily Dose: | 0.024 |
| Skin Sensitization: | 0.92 | Carcinogencity: | 0.029 |
| Eye Corrosion: | 0.299 | Eye Irritation: | 0.939 |
| Respiratory Toxicity: | 0.145 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000029 | 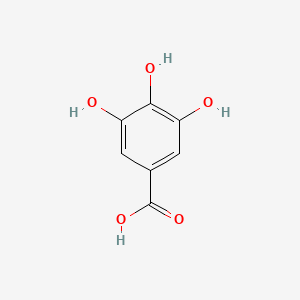 |
0.667 | D0U0OT | 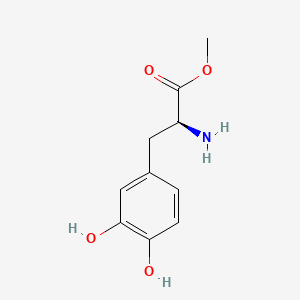 |
0.345 | ||
| ENC000729 | 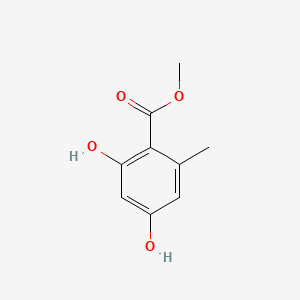 |
0.478 | D0Y7PG | 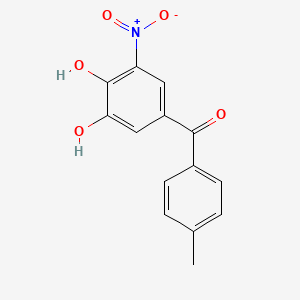 |
0.313 | ||
| ENC000002 | 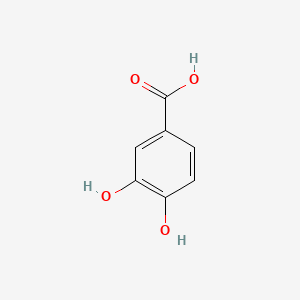 |
0.400 | D0BA6T | 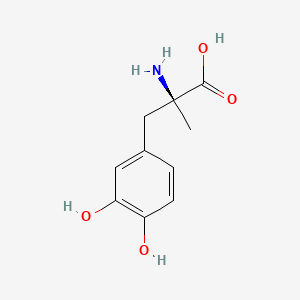 |
0.304 | ||
| ENC000674 | 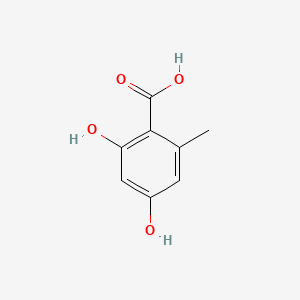 |
0.383 | D0V9EN | 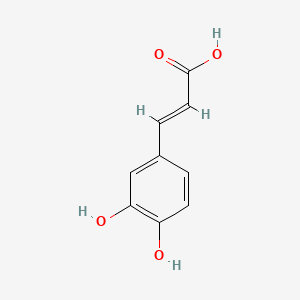 |
0.302 | ||
| ENC002095 | 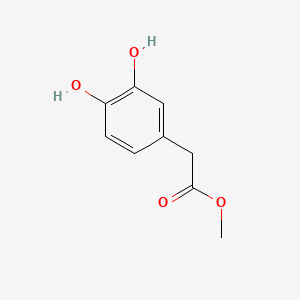 |
0.380 | D08HVR | 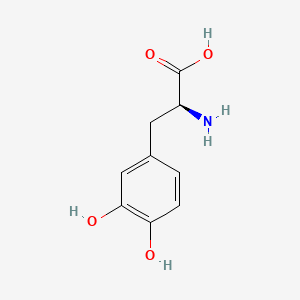 |
0.291 | ||
| ENC002875 | 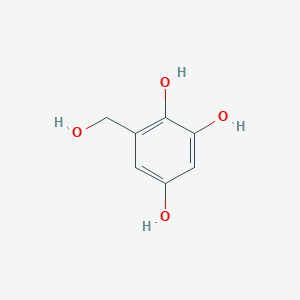 |
0.370 | D07EXH | 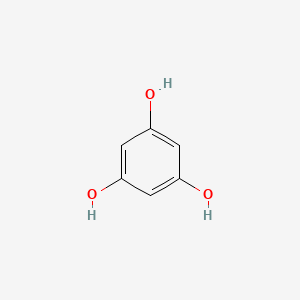 |
0.289 | ||
| ENC006073 | 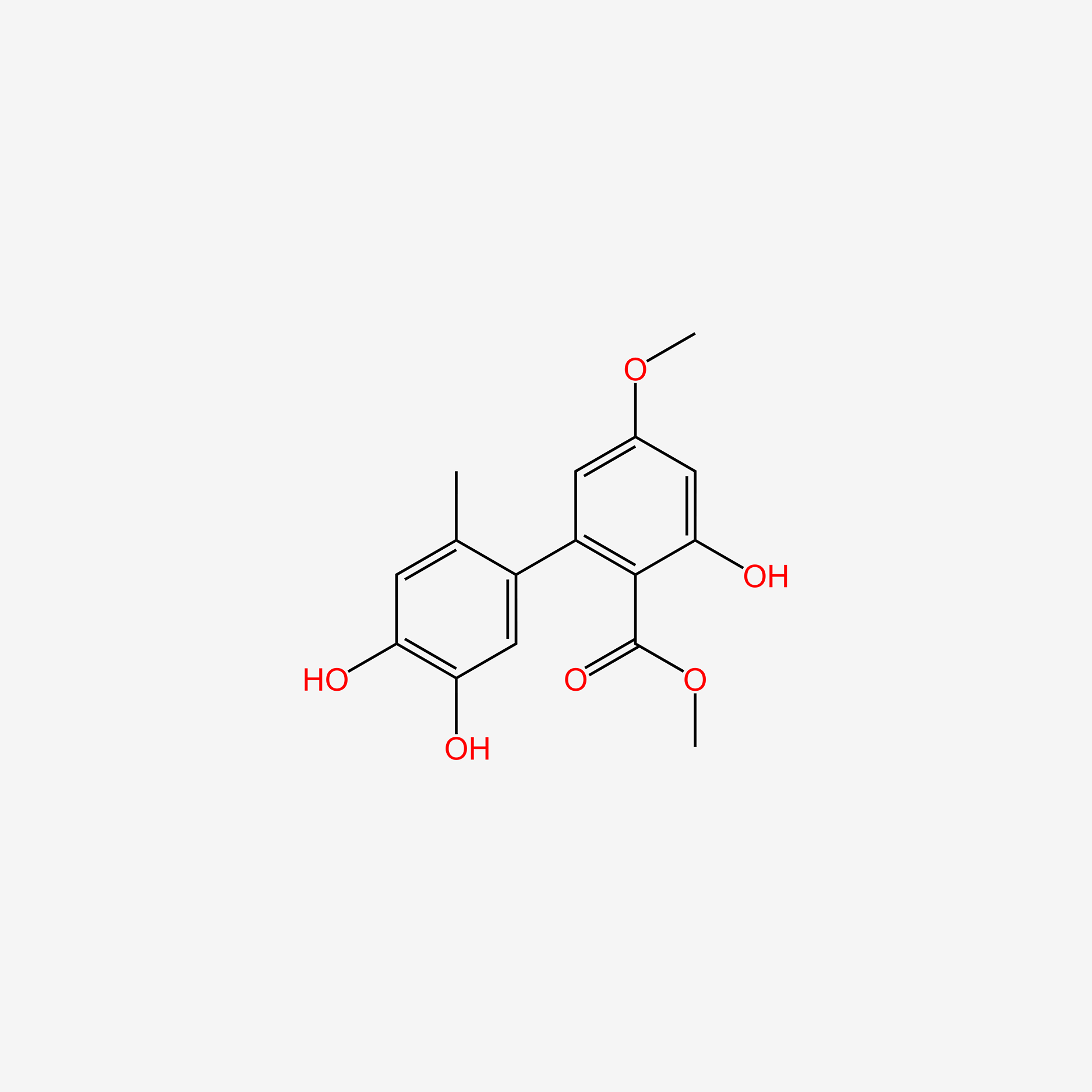 |
0.368 | D0P7JZ | 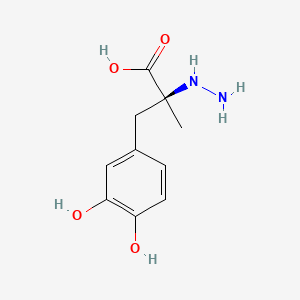 |
0.288 | ||
| ENC000367 | 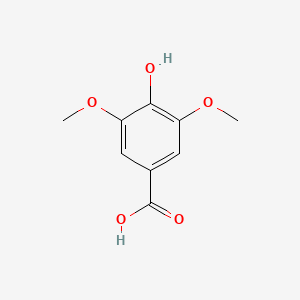 |
0.365 | D0I3RO | 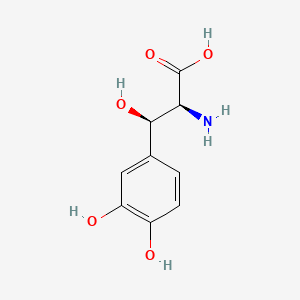 |
0.281 | ||
| ENC004830 | 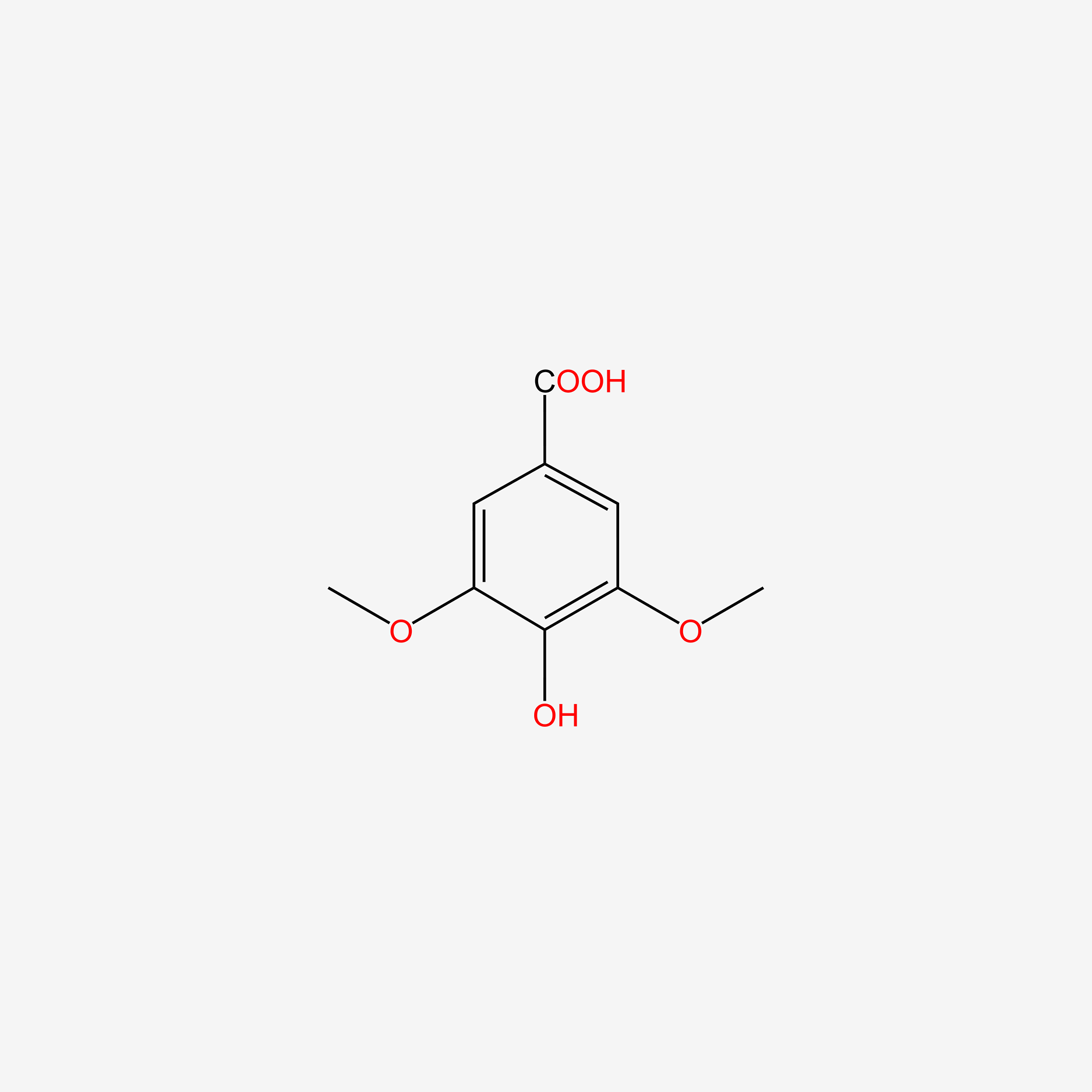 |
0.365 | D0Y6KO | 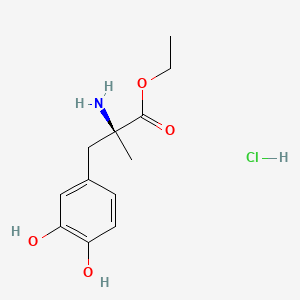 |
0.270 | ||
| ENC005900 | 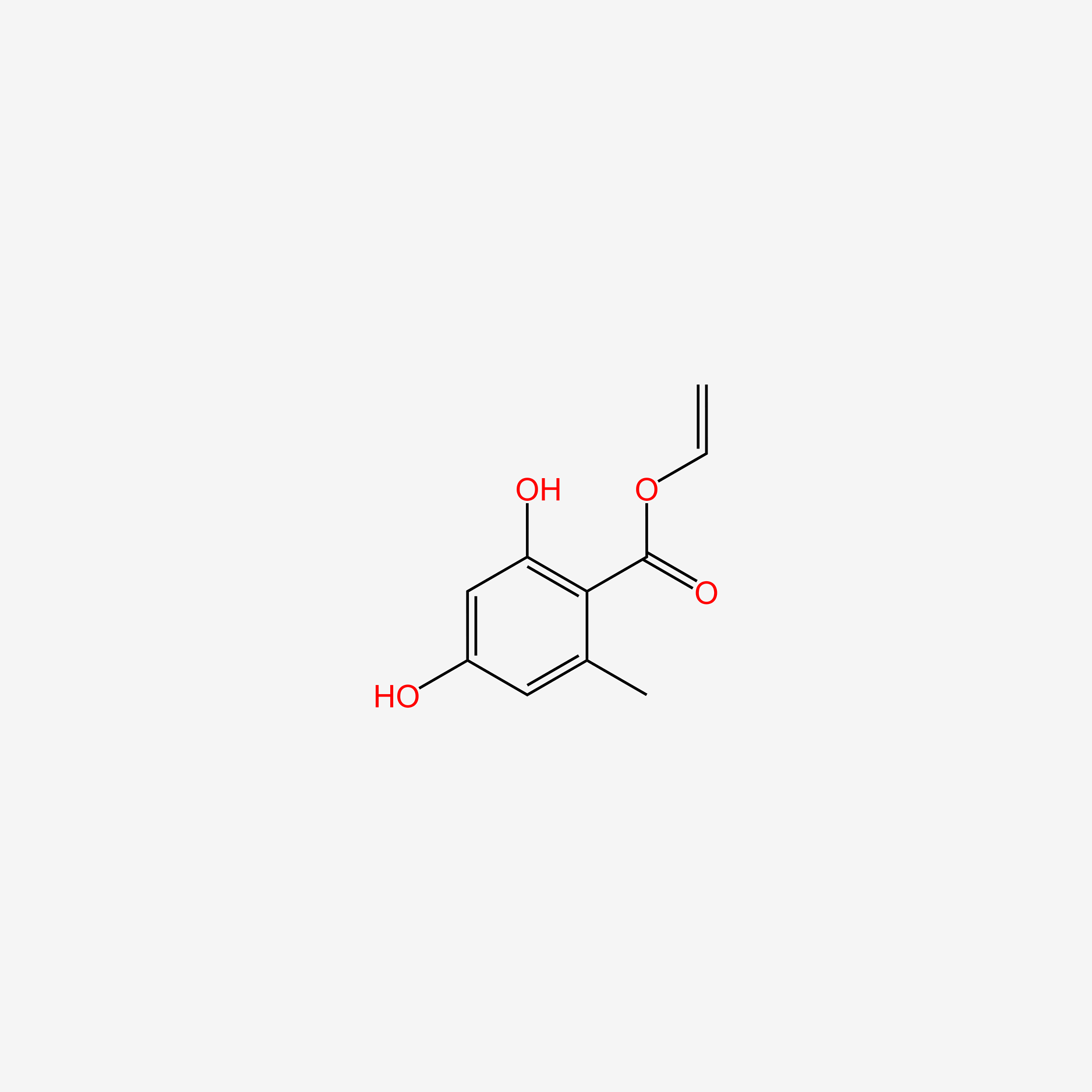 |
0.365 | D07MGA |  |
0.267 | ||