NPs Basic Information

|
Name |
Isofraxidin
|
| Molecular Formula | C11H10O5 | |
| IUPAC Name* |
7-hydroxy-6,8-dimethoxychromen-2-one
|
|
| SMILES |
COC1=C(C(=C2C(=C1)C=CC(=O)O2)OC)O
|
|
| InChI |
InChI=1S/C11H10O5/c1-14-7-5-6-3-4-8(12)16-10(6)11(15-2)9(7)13/h3-5,13H,1-2H3
|
|
| InChIKey |
HOEVRHHMDJKUMZ-UHFFFAOYSA-N
|
|
| Synonyms |
Isofraxidin; 486-21-5; Phytodolor; 7-hydroxy-6,8-dimethoxychromen-2-one; 6,8-Dimethoxyumbelliferone; 7-Hydroxy-6,8-dimethoxy-2H-1-benzopyran-2-one; 2H-1-Benzopyran-2-one, 7-hydroxy-6,8-dimethoxy-; 7-hydroxy-6,8-dimethoxy-2H-chromen-2-one; NSC-324637; CHEBI:81121; 304915F056; Umbelliferone, 6,8-dimethoxy-; Coumarin, 7-hydroxy-6,8-dimethoxy-; NSC 324637; BRN 0202652; Calycanthogenol; 6,8-dimethoxy-7-hydroxycoumarin; 7-Hydroxy-6,8-dimethoxycoumarin; Isofraxidin ,(S); UNII-304915F056; 5-18-04-00332 (Beilstein Handbook Reference); CHEMBL451518; SCHEMBL3924718; Isofraxidin, analytical standard; DTXSID70197557; HY-N0774; ZINC1573299; MFCD00221757; NSC324637; s9240; ISOFRAXIDIN B814484K143; AKOS000278010; AC-8051; CCG-266730; BS-42209; DB-050272; CS-0009797; FT-0688350; FT-0698467; C17480; 486I215; A827571; 7-hydroxy-6,8-dimethoxy-chromen-2-one;Isofraxidin; Q-100542; Q15426264
|
|
| CAS | 486-21-5 | |
| PubChem CID | 5318565 | |
| ChEMBL ID | CHEMBL451518 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 222.19 | ALogp: | 1.5 |
| HBD: | 1 | HBA: | 5 |
| Rotatable Bonds: | 2 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 65.0 | Aromatic Rings: | 2 |
| Heavy Atoms: | 16 | QED Weighted: | 0.787 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.785 | MDCK Permeability: | 0.00002360 |
| Pgp-inhibitor: | 0.024 | Pgp-substrate: | 0.002 |
| Human Intestinal Absorption (HIA): | 0.012 | 20% Bioavailability (F20%): | 0.007 |
| 30% Bioavailability (F30%): | 0.45 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.184 | Plasma Protein Binding (PPB): | 77.55% |
| Volume Distribution (VD): | 0.9 | Fu: | 19.38% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.965 | CYP1A2-substrate: | 0.963 |
| CYP2C19-inhibitor: | 0.103 | CYP2C19-substrate: | 0.332 |
| CYP2C9-inhibitor: | 0.062 | CYP2C9-substrate: | 0.768 |
| CYP2D6-inhibitor: | 0.506 | CYP2D6-substrate: | 0.692 |
| CYP3A4-inhibitor: | 0.219 | CYP3A4-substrate: | 0.297 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 12.3 | Half-life (T1/2): | 0.886 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.06 | Human Hepatotoxicity (H-HT): | 0.667 |
| Drug-inuced Liver Injury (DILI): | 0.903 | AMES Toxicity: | 0.164 |
| Rat Oral Acute Toxicity: | 0.406 | Maximum Recommended Daily Dose: | 0.055 |
| Skin Sensitization: | 0.56 | Carcinogencity: | 0.644 |
| Eye Corrosion: | 0.023 | Eye Irritation: | 0.299 |
| Respiratory Toxicity: | 0.173 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC001524 | 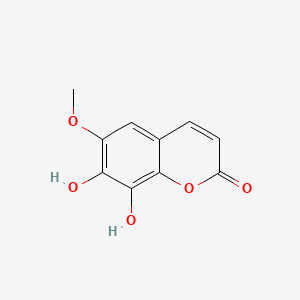 |
0.700 | D08SKH | 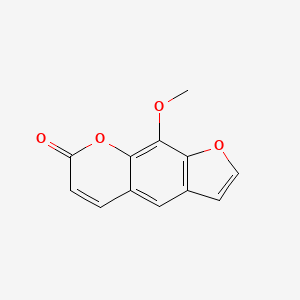 |
0.525 | ||
| ENC001537 | 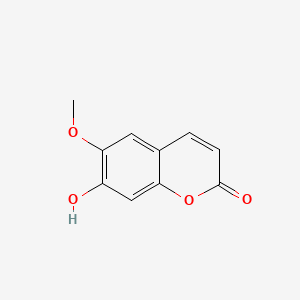 |
0.537 | D06GCK |  |
0.378 | ||
| ENC005717 |  |
0.441 | D0G4KG | 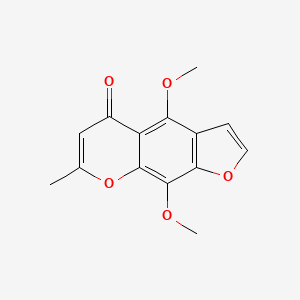 |
0.366 | ||
| ENC005716 |  |
0.441 | D0E9CD | 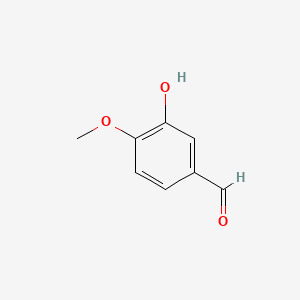 |
0.321 | ||
| ENC004990 |  |
0.429 | D09GYT | 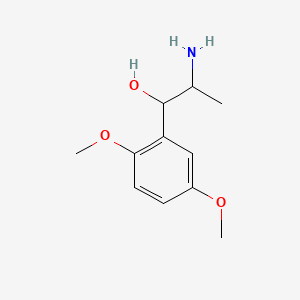 |
0.313 | ||
| ENC001472 |  |
0.412 | D0W8WB | 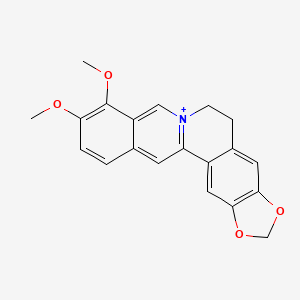 |
0.300 | ||
| ENC002897 |  |
0.408 | D02XJY | 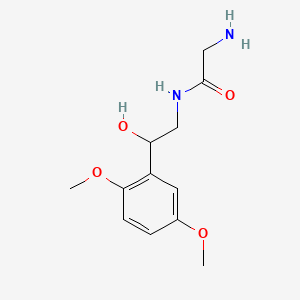 |
0.292 | ||
| ENC000982 |  |
0.397 | D0FA2O |  |
0.282 | ||
| ENC004401 |  |
0.397 | D07MGA |  |
0.280 | ||
| ENC000168 |  |
0.396 | D02LZB |  |
0.280 | ||