NPs Basic Information
|
Name |
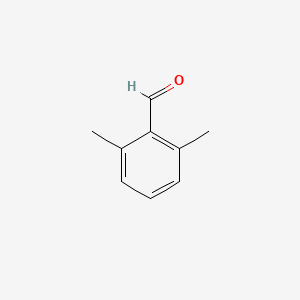
2,6-Dimethylbenzaldehyde
|
| Molecular Formula | C9H10O | |
| IUPAC Name* |
2,6-dimethylbenzaldehyde
|
|
| SMILES |
CC1=C(C(=CC=C1)C)C=O
|
|
| InChI |
InChI=1S/C9H10O/c1-7-4-3-5-8(2)9(7)6-10/h3-6H,1-2H3
|
|
| InChIKey |
QOJQBWSZHCKOLL-UHFFFAOYSA-N
|
|
| Synonyms |
2,6-Dimethylbenzaldehyde; 1123-56-4; BENZALDEHYDE, 2,6-DIMETHYL-; MFCD00128003; m-Xylene-2-carboxaldehyde; 2,6-DiMethyl-Benzaldehyde; 2,6-dimethylbenzaldehyd; 2,6-Dimethyl benzaldehyde; SCHEMBL66979; 2,6-Dimethylbenzaldehyde, 97%; DTXSID40342693; ZINC2582001; BBL103183; CL8253; STL556993; AKOS005255233; CS-W007640; PS-6110; SY002379; DB-005896; A2257; AM20050097; D3681; FT-0633367; EN300-82115; A802559; W-204781; F8889-9343; Z1233237340
|
|
| CAS | 1123-56-4 | |
| PubChem CID | 583841 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 134.17 | ALogp: | 2.1 |
| HBD: | 0 | HBA: | 1 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 17.1 | Aromatic Rings: | 1 |
| Heavy Atoms: | 10 | QED Weighted: | 0.54 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.321 | MDCK Permeability: | 0.00002670 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.061 |
| Human Intestinal Absorption (HIA): | 0.004 | 20% Bioavailability (F20%): | 0.002 |
| 30% Bioavailability (F30%): | 0.004 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.956 | Plasma Protein Binding (PPB): | 80.34% |
| Volume Distribution (VD): | 1.289 | Fu: | 20.56% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.93 | CYP1A2-substrate: | 0.89 |
| CYP2C19-inhibitor: | 0.712 | CYP2C19-substrate: | 0.804 |
| CYP2C9-inhibitor: | 0.108 | CYP2C9-substrate: | 0.734 |
| CYP2D6-inhibitor: | 0.043 | CYP2D6-substrate: | 0.867 |
| CYP3A4-inhibitor: | 0.091 | CYP3A4-substrate: | 0.334 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 5.672 | Half-life (T1/2): | 0.666 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.012 | Human Hepatotoxicity (H-HT): | 0.021 |
| Drug-inuced Liver Injury (DILI): | 0.109 | AMES Toxicity: | 0.671 |
| Rat Oral Acute Toxicity: | 0.026 | Maximum Recommended Daily Dose: | 0.569 |
| Skin Sensitization: | 0.693 | Carcinogencity: | 0.576 |
| Eye Corrosion: | 0.978 | Eye Irritation: | 0.997 |
| Respiratory Toxicity: | 0.972 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000364 |  |
0.545 | D01PJR |  |
0.422 | ||
| ENC000179 |  |
0.485 | D0X0RI |  |
0.409 | ||
| ENC000649 | 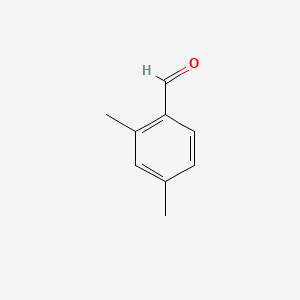 |
0.459 | D0X4RN | 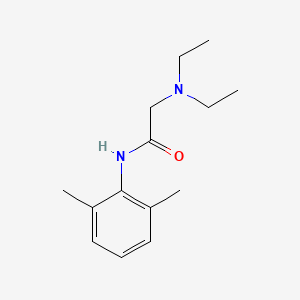 |
0.352 | ||
| ENC000552 | 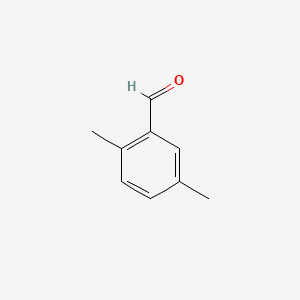 |
0.459 | D0WO8W | 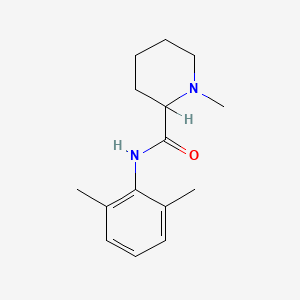 |
0.328 | ||
| ENC000414 | 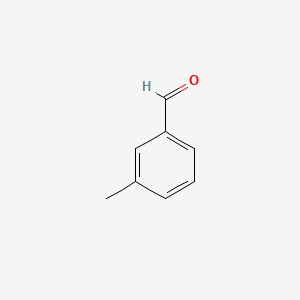 |
0.405 | D0G7DJ |  |
0.306 | ||
| ENC000180 | 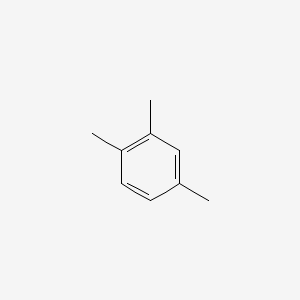 |
0.378 | D0U3DU | 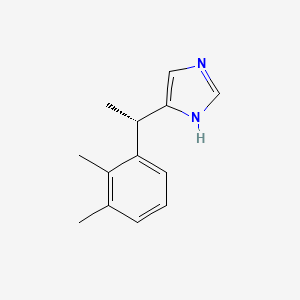 |
0.302 | ||
| ENC000614 |  |
0.378 | D09RHQ |  |
0.297 | ||
| ENC000390 | 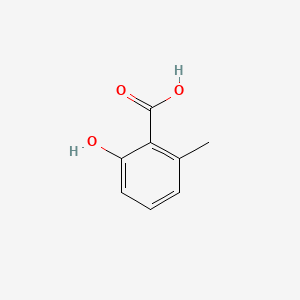 |
0.366 | D0E9CD | 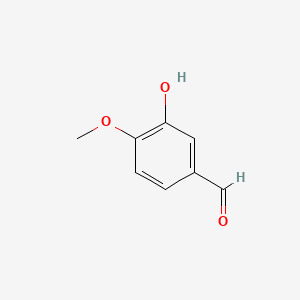 |
0.295 | ||
| ENC000239 | 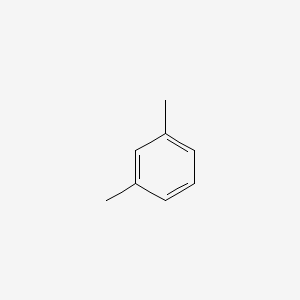 |
0.361 | D0H7AV |  |
0.292 | ||
| ENC000240 |  |
0.361 | D0A0FL | 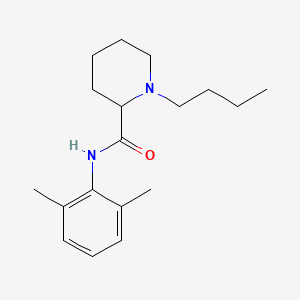 |
0.284 | ||