NPs Basic Information

|
Name |
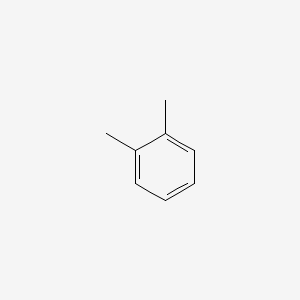
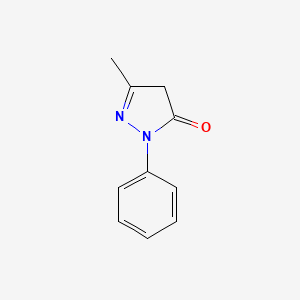
2,6-Dimethylpyridine
|
| Molecular Formula | C7H9N | |
| IUPAC Name* |
2,6-dimethylpyridine
|
|
| SMILES |
CC1=NC(=CC=C1)C
|
|
| InChI |
InChI=1S/C7H9N/c1-6-4-3-5-7(2)8-6/h3-5H,1-2H3
|
|
| InChIKey |
OISVCGZHLKNMSJ-UHFFFAOYSA-N
|
|
| Synonyms |
2,6-Dimethylpyridine; 2,6-LUTIDINE; 108-48-5; Lutidine; Pyridine, 2,6-dimethyl-; 2,6-Dimethypyridine; alpha,alpha'-Lutidine; 2,6-dimethyl-pyridine; alpha,alpha'-Dimethylpyridine; HSDB 79; FEMA No. 3540; NSC 2155; 2,6-Lutidene; .alpha.,.alpha.'-Dimethylpyridine; .alpha.,.alpha.'-Lutidine; 15FQ5D0T3P; CHEBI:32548; NSC-2155; LUT; alpha,alpha'-Lutidin; EINECS 203-587-3; MFCD00006345; lutidin; UNII-15FQ5D0T3P; AI3-24282; 2,6Lutidine; 2,6 lutidine; 2-6-lutidine; 2.6-lutidine; 2,6 -lutidine; 2,6- lutidine; 2,6-dimethylpiridine; 2.6-dimethylpyridine; 2,6-dimethyl pyridin; 2,6-dimethyl pyridine; 2,6-Lutidine, 8CI; .alpha.,.alpha.'-Lutidin; SCHEMBL9611; DSSTox_CID_30109; DSSTox_GSID_51557; CHEMBL22976; 2,6-LUTIDINE [MI]; 2,6-LUTIDINE [HSDB]; DIMETHYLPYRIDINE, 2,6-; DTXSID7051557; FEMA 3540; 2,6-Dimethylpyridine, >=99%; NSC2155; 2,6-Dimethylpyridine, redistilled; ZINC967330; 2,6-Lutidine, analytical standard; Tox21_303862; 2,6-DIMETHYLPYRIDINE [FHFI]; BBL013176; STL163956; AKOS005716680; AC-5116; AM10711; 2,6-Lutidine, ReagentPlus(R), 98%; NCGC00357127-01; BP-30085; BP-31131; CAS-108-48-5; DB-013994; FT-0610735; L0067; EN300-19113; 2,6-Lutidine, purified by redistillation, >=99%; A801883; AC-907/25014177; Q209284; Q-100049; F0001-0171; Z104472820
|
|
| CAS | 108-48-5 | |
| PubChem CID | 7937 | |
| ChEMBL ID | CHEMBL22976 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Physi-Chem Properties
| Molecular Weight: | 107.15 | ALogp: | 1.7 |
| HBD: | 0 | HBA: | 1 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 12.9 | Aromatic Rings: | 1 |
| Heavy Atoms: | 8 | QED Weighted: | 0.495 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.529 | MDCK Permeability: | 0.00003650 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.005 |
| Human Intestinal Absorption (HIA): | 0.003 | 20% Bioavailability (F20%): | 0.004 |
| 30% Bioavailability (F30%): | 0.002 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.94 | Plasma Protein Binding (PPB): | 56.68% |
| Volume Distribution (VD): | 0.818 | Fu: | 41.38% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.674 | CYP1A2-substrate: | 0.861 |
| CYP2C19-inhibitor: | 0.054 | CYP2C19-substrate: | 0.631 |
| CYP2C9-inhibitor: | 0.009 | CYP2C9-substrate: | 0.125 |
| CYP2D6-inhibitor: | 0.254 | CYP2D6-substrate: | 0.785 |
| CYP3A4-inhibitor: | 0.02 | CYP3A4-substrate: | 0.382 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 6.742 | Half-life (T1/2): | 0.447 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.026 | Human Hepatotoxicity (H-HT): | 0.265 |
| Drug-inuced Liver Injury (DILI): | 0.084 | AMES Toxicity: | 0.078 |
| Rat Oral Acute Toxicity: | 0.91 | Maximum Recommended Daily Dose: | 0.023 |
| Skin Sensitization: | 0.586 | Carcinogencity: | 0.679 |
| Eye Corrosion: | 0.963 | Eye Irritation: | 0.992 |
| Respiratory Toxicity: | 0.958 |