NPs Basic Information

|
Name |
4H-Pyran-4-one, 2,3-dihydro-3,5-dihydroxy-6-methyl-
|
| Molecular Formula | C6H8O4 | |
| IUPAC Name* |
3,5-dihydroxy-6-methyl-2,3-dihydropyran-4-one
|
|
| SMILES |
CC1=C(C(=O)C(CO1)O)O
|
|
| InChI |
InChI=1S/C6H8O4/c1-3-5(8)6(9)4(7)2-10-3/h4,7-8H,2H2,1H3
|
|
| InChIKey |
VOLMSPGWNYJHQQ-UHFFFAOYSA-N
|
|
| Synonyms |
28564-83-2; 2,3-Dihydro-3,5-dihydroxy-6-methyl-4h-pyran-4-one; 4H-Pyran-4-one, 2,3-dihydro-3,5-dihydroxy-6-methyl-; 3,5-Dihydroxy-6-methyl-2,3-dihydro-4H-pyran-4-one; 3,5-Dihydroxy-6-methyl-2H-pyran-4(3H)-one; 3,5-dihydroxy-6-methyl-2,3-dihydropyran-4-one; RAE1LU40SR; 2,3-Dihydro-3,5-dihydroxy-6-methyl-4(H)-pyran-4-one; 3,5-dihydroxy-6-methyl-3,4-dihydro-2H-pyran-4-one; dnd-omi 70 nmol/L; CCRIS 8144; UNII-RAE1LU40SR; 3-Hydroxy-2,3-dihydromaltol; SCHEMBL1278394; 4H-Pyran-4-one,2,3-dihydro-3,5-dihydroxy-6-methyl-; DTXSID00880920; AKOS006279621; DA-18707; CS-0245013; FT-0769074; 2,3-Dihydro-3,5-dihydroxy-6-methyl-4-pyrone; EN300-298133; 3,5-dihydroxy-2-me-thyl-5,6-dihydropyran-4-one; A902158; 2,3-dihydro-3,5-dihydroxy--6-methyl-4H-pyran-4-one; Z1198158588
|
|
| CAS | 28564-83-2 | |
| PubChem CID | 119838 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 144.12 | ALogp: | -0.4 |
| HBD: | 2 | HBA: | 4 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 66.8 | Aromatic Rings: | 1 |
| Heavy Atoms: | 10 | QED Weighted: | 0.508 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.812 | MDCK Permeability: | 0.00012576 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.004 |
| Human Intestinal Absorption (HIA): | 0.023 | 20% Bioavailability (F20%): | 0.005 |
| 30% Bioavailability (F30%): | 0.486 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.891 | Plasma Protein Binding (PPB): | 17.47% |
| Volume Distribution (VD): | 0.294 | Fu: | 73.50% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.017 | CYP1A2-substrate: | 0.109 |
| CYP2C19-inhibitor: | 0.029 | CYP2C19-substrate: | 0.254 |
| CYP2C9-inhibitor: | 0.012 | CYP2C9-substrate: | 0.065 |
| CYP2D6-inhibitor: | 0.004 | CYP2D6-substrate: | 0.151 |
| CYP3A4-inhibitor: | 0.005 | CYP3A4-substrate: | 0.206 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 2.761 | Half-life (T1/2): | 0.73 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.025 | Human Hepatotoxicity (H-HT): | 0.07 |
| Drug-inuced Liver Injury (DILI): | 0.869 | AMES Toxicity: | 0.913 |
| Rat Oral Acute Toxicity: | 0.233 | Maximum Recommended Daily Dose: | 0.008 |
| Skin Sensitization: | 0.426 | Carcinogencity: | 0.035 |
| Eye Corrosion: | 0.067 | Eye Irritation: | 0.837 |
| Respiratory Toxicity: | 0.591 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC005552 |  |
0.385 | D07AHW | 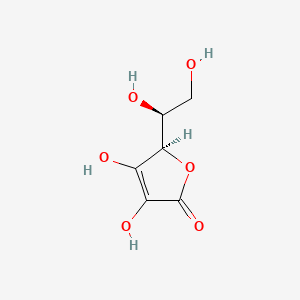 |
0.239 | ||
| ENC003178 | 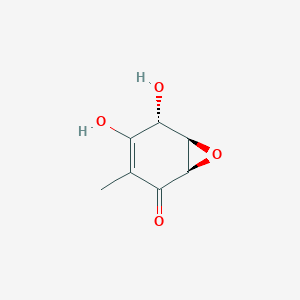 |
0.341 | D0CL9S | 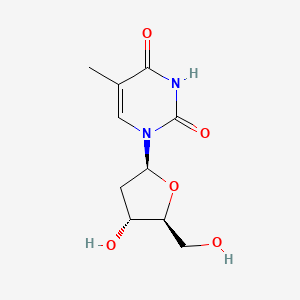 |
0.220 | ||
| ENC000788 | 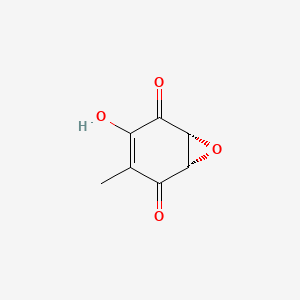 |
0.310 | D0N0OU | 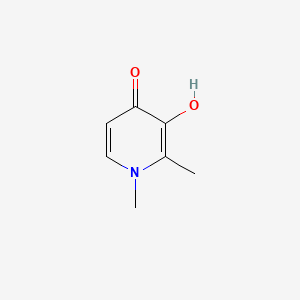 |
0.209 | ||
| ENC003046 | 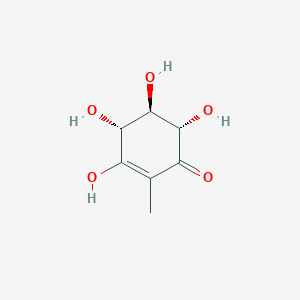 |
0.302 | D04VIS | 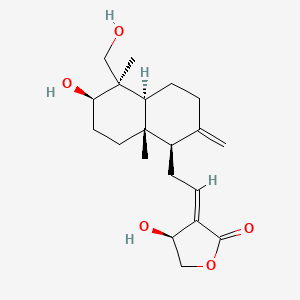 |
0.205 | ||
| ENC004403 |  |
0.280 | D0Z8AA | 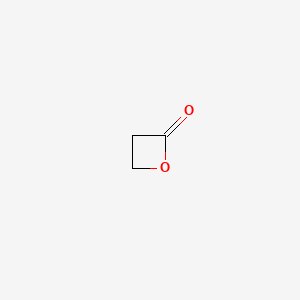 |
0.188 | ||
| ENC006048 |  |
0.280 | D0R2KF |  |
0.185 | ||
| ENC006049 |  |
0.280 | D02WFK |  |
0.184 | ||
| ENC005580 |  |
0.278 | D03KXY |  |
0.183 | ||
| ENC003894 |  |
0.263 | D0TS1Z |  |
0.180 | ||
| ENC005308 |  |
0.263 | D09PZO |  |
0.180 | ||