NPs Basic Information
|
Name |
2-Ethyl-3-methylpyrazine
|
| Molecular Formula | C7H10N2 | |
| IUPAC Name* |
2-ethyl-3-methylpyrazine
|
|
| SMILES |
CCC1=NC=CN=C1C
|
|
| InChI |
InChI=1S/C7H10N2/c1-3-7-6(2)8-4-5-9-7/h4-5H,3H2,1-2H3
|
|
| InChIKey |
LNIMMWYNSBZESE-UHFFFAOYSA-N
|
|
| Synonyms |
2-ETHYL-3-METHYLPYRAZINE; 15707-23-0; Pyrazine, 2-ethyl-3-methyl-; 3-Ethyl-2-methylpyrazine; 2-Methyl-3-ethylpyrazine; 2-Ethyl-3-methyl pyrazine; Pyrazine, ethylmethyl-; 2-ethyl-3-methyl-pyrazine; FEMA No. 3155; 2-Ethyl-3-methylpyrazine, 9CI; Pyrazine, 3-ethyl-2-methyl; 9GF35MK66U; BRN 0956775; EINECS 239-799-8; 3-ethyl-2-methyl pyrazine; UNII-9GF35MK66U; filbert pyrazine; 2-Methyl-3-ethyl pyrazine; 2-Methyl-3-ethyl-pyrazine; DSSTox_CID_27464; DSSTox_RID_82364; DSSTox_GSID_47464; 5-23-05-00418 (Beilstein Handbook Reference); SCHEMBL108460; CHEMBL3187840; DTXSID4047464; FEMA 3155; CHEBI:193620; ZINC409274; AMY23218; Tox21_302613; MFCD00006150; AKOS015842908; CS-W013548; 2-ETHYL-3-METHYL-1,4-PYRAZINE; 2-ETHYL-3-METHYLPYRAZINE [FCC]; 2-ETHYL-3-METHYLPYRAZINE [FHFI]; NCGC00256746-01; AC-16592; BS-15466; CAS-15707-23-0; DB-021037; E0361; FT-0612243; 2-Ethyl-3-methylpyrazine, >=98%, FCC, FG; D90495; EN300-2010184; A809805; Q-100188; Q17239258; 2-Ethyl-3-methylpyrazine, analytical reference material; 2-Methyl-5(or 3)-ethylpyrazine; 2-Methyl-5(or 3)-ethyl -1,4-diazine
|
|
| CAS | 15707-23-0 | |
| PubChem CID | 27457 | |
| ChEMBL ID | CHEMBL3187840 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 122.17 | ALogp: | 1.1 |
| HBD: | 0 | HBA: | 2 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 25.8 | Aromatic Rings: | 1 |
| Heavy Atoms: | 9 | QED Weighted: | 0.568 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.32 | MDCK Permeability: | 0.00003320 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.003 |
| Human Intestinal Absorption (HIA): | 0.004 | 20% Bioavailability (F20%): | 0.006 |
| 30% Bioavailability (F30%): | 0.028 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.834 | Plasma Protein Binding (PPB): | 27.99% |
| Volume Distribution (VD): | 1.591 | Fu: | 71.50% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.2 | CYP1A2-substrate: | 0.666 |
| CYP2C19-inhibitor: | 0.04 | CYP2C19-substrate: | 0.8 |
| CYP2C9-inhibitor: | 0.006 | CYP2C9-substrate: | 0.511 |
| CYP2D6-inhibitor: | 0.009 | CYP2D6-substrate: | 0.668 |
| CYP3A4-inhibitor: | 0.016 | CYP3A4-substrate: | 0.304 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 5.075 | Half-life (T1/2): | 0.494 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.014 | Human Hepatotoxicity (H-HT): | 0.364 |
| Drug-inuced Liver Injury (DILI): | 0.769 | AMES Toxicity: | 0.039 |
| Rat Oral Acute Toxicity: | 0.479 | Maximum Recommended Daily Dose: | 0.175 |
| Skin Sensitization: | 0.864 | Carcinogencity: | 0.208 |
| Eye Corrosion: | 0.744 | Eye Irritation: | 0.991 |
| Respiratory Toxicity: | 0.848 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
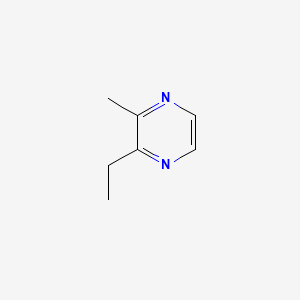
| ENC000650 | 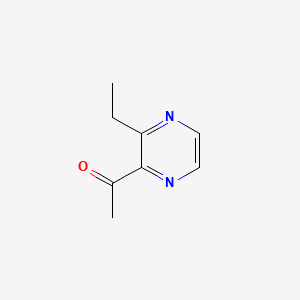 |
0.571 | D0P0HB | 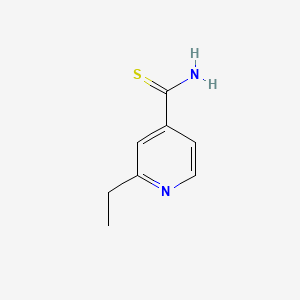 |
0.250 | ||
| ENC000599 | 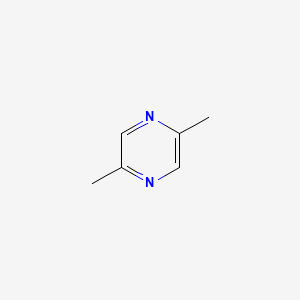 |
0.306 | D0XF8W |  |
0.220 | ||
| ENC000657 |  |
0.257 | D0U2CV |  |
0.218 | ||
| ENC000413 | 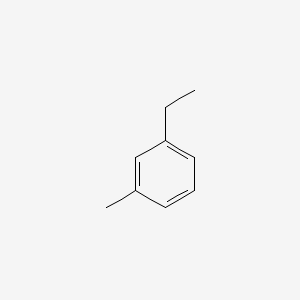 |
0.250 | D0L7UQ | 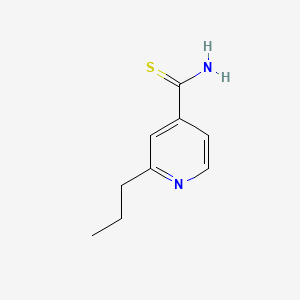 |
0.208 | ||
| ENC000407 | 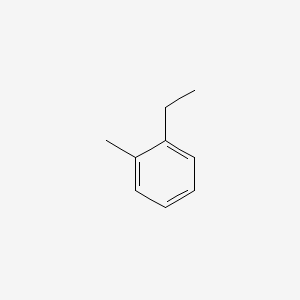 |
0.250 | D0C0SK |  |
0.200 | ||
| ENC000498 | 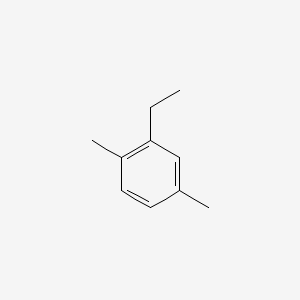 |
0.238 | D0LM4A | 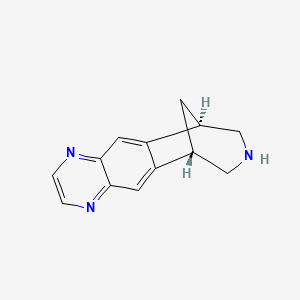 |
0.197 | ||
| ENC000734 | 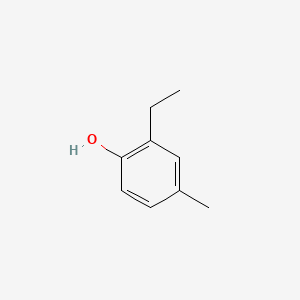 |
0.238 | D0AE3X | 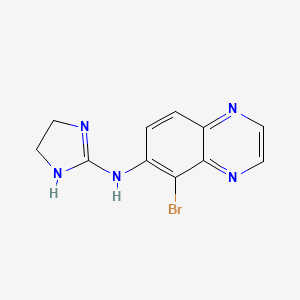 |
0.190 | ||
| ENC001061 | 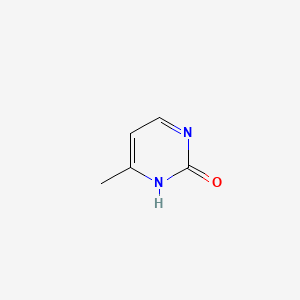 |
0.237 | D07MUN |  |
0.184 | ||
| ENC000648 |  |
0.231 | D0V5IW | 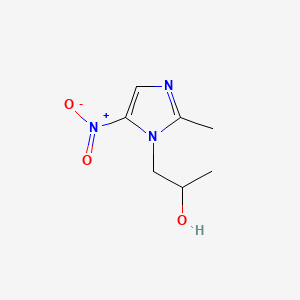 |
0.180 | ||
| ENC001026 | 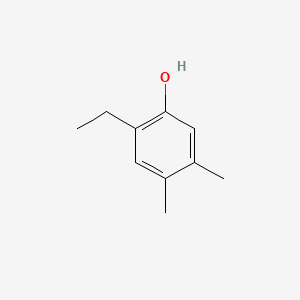 |
0.227 | D0O4SE |  |
0.175 | ||