NPs Basic Information
|
Name |
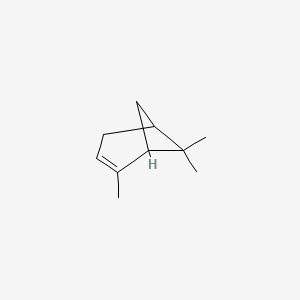
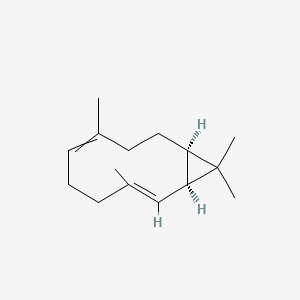
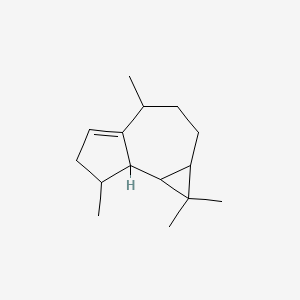
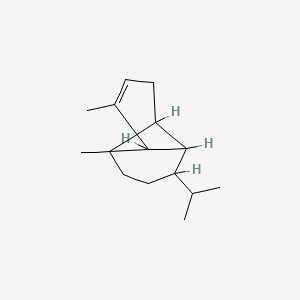
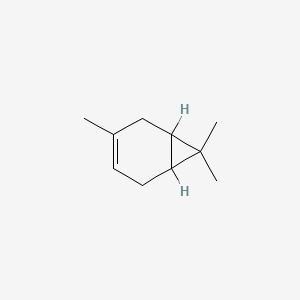
3-Carene
|
| Molecular Formula | C10H16 | |
| IUPAC Name* |
3,7,7-trimethylbicyclo[4.1.0]hept-3-ene
|
|
| SMILES |
CC1=CCC2C(C1)C2(C)C
|
|
| InChI |
InChI=1S/C10H16/c1-7-4-5-8-9(6-7)10(8,2)3/h4,8-9H,5-6H2,1-3H3
|
|
| InChIKey |
BQOFWKZOCNGFEC-UHFFFAOYSA-N
|
|
| Synonyms |
3-Carene; 13466-78-9; 3,7,7-Trimethylbicyclo[4.1.0]hept-3-ene; Delta-3-Carene; Car-3-ene; Carene; Bicyclo[4.1.0]hept-3-ene, 3,7,7-trimethyl-; Delta-car-3-ene; (+-)-delta3-Carene; (+-)-3-Carene; CHEBI:35661; Bicyclo(4.1.0)hept-3-ene, 3,7,7(or 4,7,7)-trimethyl-; 3,7,7-Trimethylbicyclo[4.1.0]-3-heptene; 3,7,7-trimethyl-bicyclo[4.1.0]hept-3-ene; 74806-04-5; Bicyclo[4.1.0]hept-3-ene, 3,7,7(or 4,7,7)-trimethyl-; (+)Car-3-ene; .delta. 3-carene; 3-.delta.-Carene; DELTA3-Carene; .DELTA.-caR-3-ene; 4,7,7-Trimethyl-3-norcarene; alpha-Carene; 3,7,7-trimethyl bicyclohept-3-ene; 4,7,7-trimethylbicyclo[4.1.0]hept-3-ene; 3-delta-Carene; Delta(3)-Carene; carene (delta-3-); 3,7,7(or 4,7,7)-Trimethylbicyclo(4.1.0)hept-3-ene; delta-3-Carene (GC); 3-Carene, 90%; Delta 3 Carene 90 PF; 3-Carene, >=90%; Bicyclo[4.1.0]hept-3-ene, 3,7,7-trimethyl-, (1S)-; DSSTox_CID_27462; DSSTox_RID_82362; DSSTox_GSID_47462; 3-Carene, analytical standard; CHEMBL506854; DTXSID4047462; HY-N6663; Tox21_302632; MFCD00001315; s5595; AKOS015840953; CCG-266136; NCGC00256842-01; AS-80902; CAS-13466-78-9; DB-063033; CS-0083202; FT-0624500; FT-0651899; E77192; EN300-173315; W-110341
|
|
| CAS | 13466-78-9 | |
| PubChem CID | 26049 | |
| ChEMBL ID | CHEMBL506854 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Physi-Chem Properties
| Molecular Weight: | 136.23 | ALogp: | 2.8 |
| HBD: | 0 | HBA: | 0 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 0.0 | Aromatic Rings: | 2 |
| Heavy Atoms: | 10 | QED Weighted: | 0.444 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.307 | MDCK Permeability: | 0.00002100 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0 |
| Human Intestinal Absorption (HIA): | 0.003 | 20% Bioavailability (F20%): | 0.125 |
| 30% Bioavailability (F30%): | 0.127 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.804 | Plasma Protein Binding (PPB): | 91.52% |
| Volume Distribution (VD): | 3.601 | Fu: | 11.94% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.706 | CYP1A2-substrate: | 0.324 |
| CYP2C19-inhibitor: | 0.339 | CYP2C19-substrate: | 0.839 |
| CYP2C9-inhibitor: | 0.275 | CYP2C9-substrate: | 0.77 |
| CYP2D6-inhibitor: | 0.013 | CYP2D6-substrate: | 0.448 |
| CYP3A4-inhibitor: | 0.045 | CYP3A4-substrate: | 0.23 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 16.061 | Half-life (T1/2): | 0.132 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.01 | Human Hepatotoxicity (H-HT): | 0.172 |
| Drug-inuced Liver Injury (DILI): | 0.049 | AMES Toxicity: | 0.004 |
| Rat Oral Acute Toxicity: | 0.035 | Maximum Recommended Daily Dose: | 0.155 |
| Skin Sensitization: | 0.785 | Carcinogencity: | 0.147 |
| Eye Corrosion: | 0.941 | Eye Irritation: | 0.98 |
| Respiratory Toxicity: | 0.266 |