NPs Basic Information

|
Name |
1-Methylpyrrole-2-carboxaldehyde
|
| Molecular Formula | C6H7NO | |
| IUPAC Name* |
1-methylpyrrole-2-carbaldehyde
|
|
| SMILES |
CN1C=CC=C1C=O
|
|
| InChI |
InChI=1S/C6H7NO/c1-7-4-2-3-6(7)5-8/h2-5H,1H3
|
|
| InChIKey |
OUKQTRFCDKSEPL-UHFFFAOYSA-N
|
|
| Synonyms |
1192-58-1; 1-Methylpyrrole-2-carboxaldehyde; 1-Methyl-1H-pyrrole-2-carbaldehyde; N-Methylpyrrole-2-carboxaldehyde; 1-Methylpyrrole-2-carbaldehyde; N-Methyl-2-pyrrolecarboxaldehyde; 1-Methyl-1H-pyrrole-2-carboxaldehyde; 2-Formyl-1-methylpyrrole; 1-Methyl-2-pyrrolecarboxaldehyde; 1H-Pyrrole-2-carboxaldehyde, 1-methyl-; 1-Methyl-2-formylpyrrole; NSC 72386; 1H-Pyrrolecarboxaldehyde, 1-methyl-; M0HYH3D7SX; 1-Methyl-2-pyrrolaldehyde; N-methylpyrrole-2-carbaldehyde; N-METHYL-2-FORMYLPYRROLE; 1-methylpyrrole-2-carboxyaldehyde; N-Methylpyrrole-2-carboxy aldehyde; NSC-72386; EINECS 214-755-0; UNII-M0HYH3D7SX; 1-Methylpyrrole-2-aldehyde; BRN 0107811; 1-methylpyrrol-2-carbaldehyde; NSC72386; MFCD00003087; 1-Methylformylpyrrole; N-Methylpyrrole-2-aldehyde; WLN: T5NJ A1 BVH; 5-21-07-00177 (Beilstein Handbook Reference); SCHEMBL260478; n-methyl-2-pyrrolecarbaldehyde; 1methyl-2-pyrrolecarboxaldehyde; N-methylpyrrol-2-carboxaldehyde; 1-Methyl-2-formyl-1H-pyrrole; CHEMBL2229659; FEMA NO. 4332; DTXSID20152338; methyl-1H-pyrrole-2-carbaldehyde; 1-methyl pyrrole-2-carboxaldehyde; 1-methyl-2-pyrrole carboxaldehyde; CHEBI:193607; 1-Methyl-Pyrrole-2-carboxaldehyde; ZINC130187; 1-methyl-1H-pyrrol-2-carbaldehyde; AMY19098; BBL001423; Pyrrole-2-carboxaldehyde, 1-methyl-; STK802640; AKOS000113751; CS-W021362; DS-1305; SB62009; N-Methyl-2-pyrrolecarboxaldehyde, 98%; FT-0608096; M1119; EN300-20969; P15926; W-108523; 1-METHYL-1H-PYRROLE-2-CARBOXALDEHYDE [FHFI]; N-Methyl-2-pyrrolecarboxaldehyde, analytical standard; Q27283313; F2190-0578; Z104485562
|
|
| CAS | 1192-58-1 | |
| PubChem CID | 14504 | |
| ChEMBL ID | CHEMBL2229659 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 109.13 | ALogp: | 0.5 |
| HBD: | 0 | HBA: | 1 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 22.0 | Aromatic Rings: | 1 |
| Heavy Atoms: | 8 | QED Weighted: | 0.497 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.524 | MDCK Permeability: | 0.00002350 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.003 |
| Human Intestinal Absorption (HIA): | 0.053 | 20% Bioavailability (F20%): | 0.004 |
| 30% Bioavailability (F30%): | 0.01 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.897 | Plasma Protein Binding (PPB): | 23.51% |
| Volume Distribution (VD): | 1.724 | Fu: | 76.75% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.897 | CYP1A2-substrate: | 0.757 |
| CYP2C19-inhibitor: | 0.099 | CYP2C19-substrate: | 0.53 |
| CYP2C9-inhibitor: | 0.015 | CYP2C9-substrate: | 0.262 |
| CYP2D6-inhibitor: | 0.003 | CYP2D6-substrate: | 0.501 |
| CYP3A4-inhibitor: | 0.008 | CYP3A4-substrate: | 0.246 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 9.187 | Half-life (T1/2): | 0.839 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.007 | Human Hepatotoxicity (H-HT): | 0.102 |
| Drug-inuced Liver Injury (DILI): | 0.039 | AMES Toxicity: | 0.05 |
| Rat Oral Acute Toxicity: | 0.075 | Maximum Recommended Daily Dose: | 0.115 |
| Skin Sensitization: | 0.105 | Carcinogencity: | 0.224 |
| Eye Corrosion: | 0.438 | Eye Irritation: | 0.893 |
| Respiratory Toxicity: | 0.783 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000640 |  |
0.438 | D0N0OU | 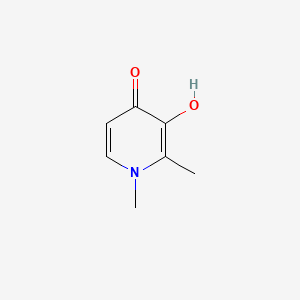 |
0.263 | ||
| ENC000190 | 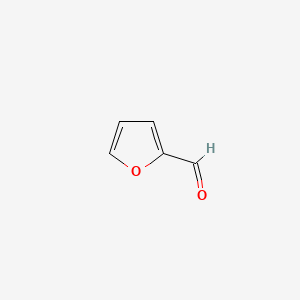 |
0.313 | D0S4BR | 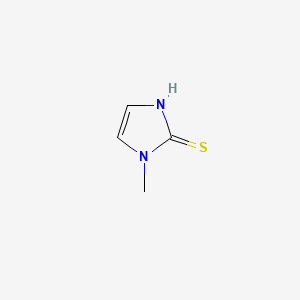 |
0.242 | ||
| ENC000414 | 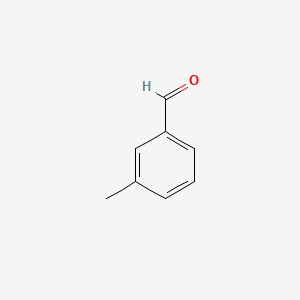 |
0.306 | D0E9CD | 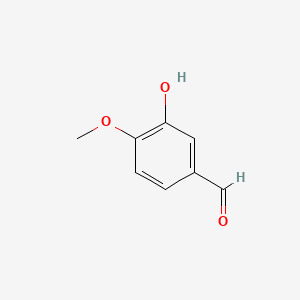 |
0.238 | ||
| ENC001334 | 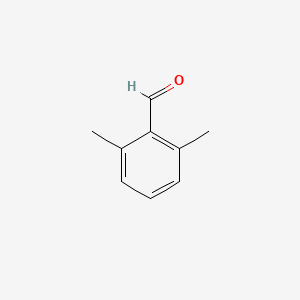 |
0.289 | D0TY5N | 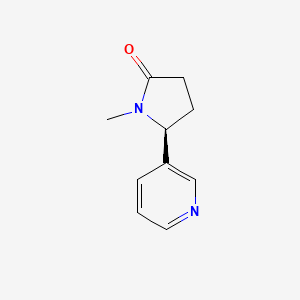 |
0.204 | ||
| ENC000012 | 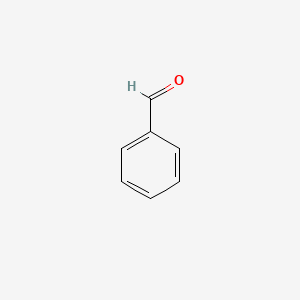 |
0.286 | D0O7JW | 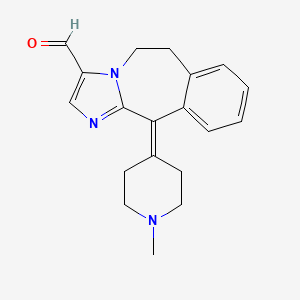 |
0.203 | ||
| ENC000166 |  |
0.270 | D02CKX | 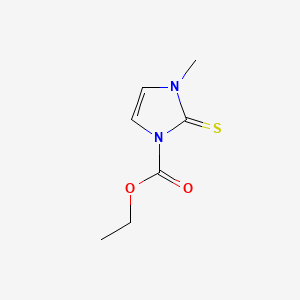 |
0.200 | ||
| ENC000412 |  |
0.257 | D06BYV | 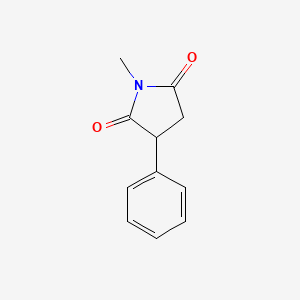 |
0.196 | ||
| ENC000552 | 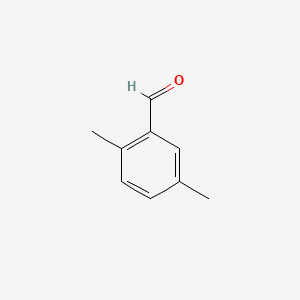 |
0.256 | D08EOD | 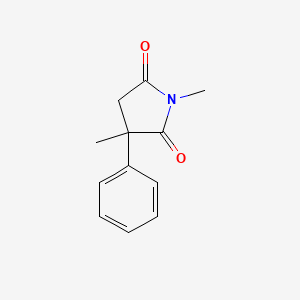 |
0.189 | ||
| ENC000649 | 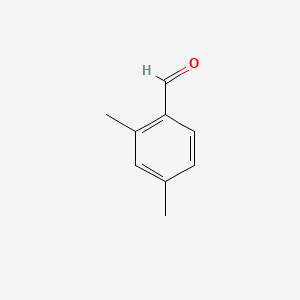 |
0.256 | D05QIM | 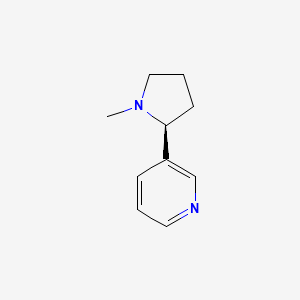 |
0.188 | ||
| ENC001839 |  |
0.250 | D0X7NU | 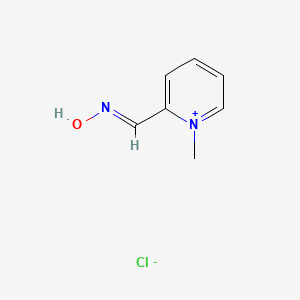 |
0.186 | ||