NPs Basic Information

|
Name |
5-Methylfurfural
|
| Molecular Formula | C6H6O2 | |
| IUPAC Name* |
5-methylfuran-2-carbaldehyde
|
|
| SMILES |
CC1=CC=C(O1)C=O
|
|
| InChI |
InChI=1S/C6H6O2/c1-5-2-3-6(4-7)8-5/h2-4H,1H3
|
|
| InChIKey |
OUDFNZMQXZILJD-UHFFFAOYSA-N
|
|
| Synonyms |
5-METHYLFURFURAL; 620-02-0; 5-Methyl-2-furaldehyde; 5-Methylfuran-2-carbaldehyde; 5-Methyl furfural; 5-Methyl-2-furfural; 2-Furancarboxaldehyde, 5-methyl-; 5-Methylfuran-2-al; 5-Methyl-2-furancarboxaldehyde; 5-Methylfurfuraldehyde; 2-Formyl-5-methylfuran; 2-Methyl-5-formylfuran; 5-Methyl-2-furfuraldehyde; 2-Furaldehyde, 5-methyl-; FEMA No. 2702; 5-methyl-furfural; 5-methyl-2-furancarbaldehyde; 5-methyl-furan-2-carbaldehyde; 5-Methyl-2-furanaldehyde; MFCD00003232; CHEBI:2091; 4482BZC72D; CCRIS 2921; EINECS 210-622-6; BRN 0106895; UNII-4482BZC72D; AI3-36591; 5-MethyIfurfural; 5-METHYLFURAN-2-ALDEHYDE; Methyl-5-furfural; alpha-Methylfurfural; 5-methyl furaldehyde; Furfural, 5-methyl-; 5-methyl furfuraldehyde; 2-methylfuran-5-aldehyde; METHYL-5-FURALDEHYDE; SCHEMBL51606; ...5-Methyl-2-furaldehyde; 5-methylfuran-2 carbaldehyde; METHYL FURFURAL, 5-; 5-17-09-00404 (Beilstein Handbook Reference); 5-methylfuran-2-carboxaldehyde; QSPL 010; 5-methyl-2-furancarboxyaldehyde; CHEMBL2230304; DTXSID1060714; SCHEMBL11096252; FEMA 2702; OUDFNZMQXZILJD-UHFFFAOYSA-; 5-METHYL FURFURAL [FCC]; 5-METHYL FURFURAL [FHFI]; 5-Methylfurfural, >=98%, FG; ZINC900814; 5-METHYL-2-FURFURYLALDEHYDE; AMY23296; BCP31055; HY-Y0543; STR02593; AC7802; GEO-01838; s9381; AKOS000119751; CCG-266047; CS-W020103; 2-FORMYL-5-METHYLTETRAHYDROFURAN; 5-Methylfurfural, ReagentPlus(R), 99%; AC-34339; BP-30242; SY001535; FT-0620652; M0254; EN300-17263; 5-Methylfurfural, analytical reference material; 5-Methylfurfural, Vetec(TM) reagent grade, 98%; A833524; 5-Methylfuran-2-carbaldehyde;5-Methyl-2-furaldehyde; J-640041; Q-100716; Q22830264; Z56899216; F2190-0577
|
|
| CAS | 620-02-0 | |
| PubChem CID | 12097 | |
| ChEMBL ID | CHEMBL2230304 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 110.11 | ALogp: | 0.7 |
| HBD: | 0 | HBA: | 2 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 30.2 | Aromatic Rings: | 1 |
| Heavy Atoms: | 8 | QED Weighted: | 0.517 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.532 | MDCK Permeability: | 0.00002050 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.753 |
| Human Intestinal Absorption (HIA): | 0.005 | 20% Bioavailability (F20%): | 0.026 |
| 30% Bioavailability (F30%): | 0.017 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.919 | Plasma Protein Binding (PPB): | 70.80% |
| Volume Distribution (VD): | 1.703 | Fu: | 51.47% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.906 | CYP1A2-substrate: | 0.846 |
| CYP2C19-inhibitor: | 0.301 | CYP2C19-substrate: | 0.406 |
| CYP2C9-inhibitor: | 0.038 | CYP2C9-substrate: | 0.736 |
| CYP2D6-inhibitor: | 0.005 | CYP2D6-substrate: | 0.709 |
| CYP3A4-inhibitor: | 0.01 | CYP3A4-substrate: | 0.333 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 6.582 | Half-life (T1/2): | 0.727 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.026 | Human Hepatotoxicity (H-HT): | 0.064 |
| Drug-inuced Liver Injury (DILI): | 0.228 | AMES Toxicity: | 0.374 |
| Rat Oral Acute Toxicity: | 0.122 | Maximum Recommended Daily Dose: | 0.043 |
| Skin Sensitization: | 0.134 | Carcinogencity: | 0.888 |
| Eye Corrosion: | 0.962 | Eye Irritation: | 0.992 |
| Respiratory Toxicity: | 0.473 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC001019 |  |
0.516 | D0E9CD | 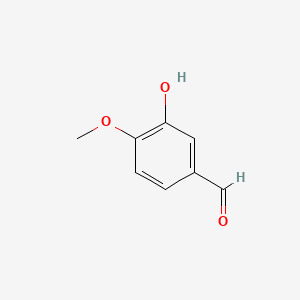 |
0.268 | ||
| ENC004043 |  |
0.415 | D0IE1E |  |
0.250 | ||
| ENC004044 |  |
0.415 | D03CUF | 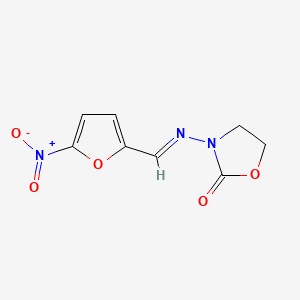 |
0.218 | ||
| ENC000190 | 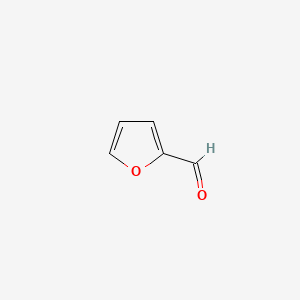 |
0.355 | D0FC1J |  |
0.213 | ||
| ENC000414 | 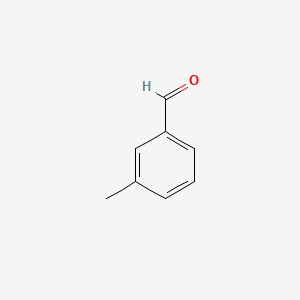 |
0.343 | D0R0BX |  |
0.211 | ||
| ENC000552 |  |
0.324 | D08ZEB |  |
0.205 | ||
| ENC000649 |  |
0.324 | D0N0OU |  |
0.200 | ||
| ENC001334 | 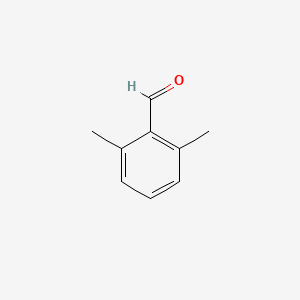 |
0.324 | D0R9OH |  |
0.190 | ||
| ENC002801 |  |
0.278 | D0T3NY | 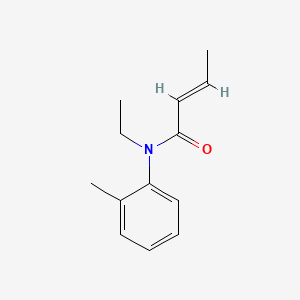 |
0.189 | ||
| ENC002238 |  |
0.270 | D06GIP | 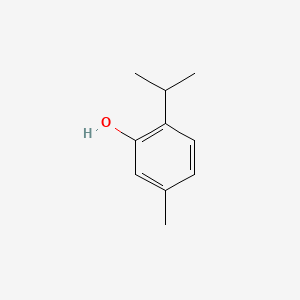 |
0.186 | ||