NPs Basic Information

|
Name |
N,N-Dimethyldodecylamine
|
| Molecular Formula | C14H31N | |
| IUPAC Name* |
N,N-dimethyldodecan-1-amine
|
|
| SMILES |
CCCCCCCCCCCCN(C)C
|
|
| InChI |
InChI=1S/C14H31N/c1-4-5-6-7-8-9-10-11-12-13-14-15(2)3/h4-14H2,1-3H3
|
|
| InChIKey |
YWFWDNVOPHGWMX-UHFFFAOYSA-N
|
|
| Synonyms |
N,N-Dimethyldodecylamine; 112-18-5; N,N-dimethyldodecan-1-amine; Dodecyldimethylamine; Dimethyl lauramine; Lauryldimethylamine; Antioxidant DDA; N,N-Dimethyl-n-dodecylamine; 1-Dodecanamine, N,N-dimethyl-; N,N-Dimethyllaurylamine; DDA (antioxidant); Barlene 125; N-Lauryldimethylamine; N-Dodecyldimethylamine; Dimethyl-n-dodecylamine; Empigen AB; Monolauryl dimethylamine; DDA (corrosion inhibitor); Armeen DM-12D; Farmin DM 20; Genamin LA 302D; Dodecylamine, N,N-dimethyl-; Farmin DM 2098; Dimethyldodecylamine; ADMA 2; Lauryl dimethyl amine; Armeen DM 12D; RC 5629; 1-(Dimethylamino)dodecane; N,N-DIMETHYL-1-DODECANAMINE; NSC 7332; NSC-7332; 6V2OM30I1Z; CHEMBL109737; 68391-04-8; Dimethyl laurylamine; Barlene 12S; Dodecyl dimethyl amine; HSDB 5568; Adma 12; EINECS 203-943-8; UNII-6V2OM30I1Z; AI3-16726; lauryl dimethylamine; Onamine 12; dimethyldodecyl amine; dodecyl dimethylamine; dodecyldimethyl amine; EINECS 269-923-6; MFCD00008970; Dodecyl-dimethyl-amine; Kemamine T-6902; 1-Dimethylaminododecane; Dodecylamine,N-dimethyl-; SDA 16-040-00; N,N-dimethyl-dodecylamine; dimethylmono-n-dodecylamine; DSSTox_CID_6906; N,N-dimethyl-1-dodecamine; EC 203-943-8; EC 269-923-6; 1,1-Dimethyl-aminododecane; 1-Dodecanamine,N-dimethyl-; DSSTox_RID_78247; DSSTox_GSID_26906; N-Dodecyl-N,N-dimethylamine; SCHEMBL107058; DTXSID1026906; N,N-Dimethyldodecylamine, 97%; DIMETHYL LAURAMINE [INCI]; NSC7332; ZINC1687260; EINECS 269-915-2; Tox21_303073; BBL011370; BDBM50147570; STL146467; AKOS005720939; WLN: 12N1 & 1; NCGC00164121-01; NCGC00257196-01; CAS-112-18-5; VS-02931; DB-041048; N,N-DIMETHYL-1-DODECANAMINE [HSDB]; CS-0297531; D0002; FT-0629557; FT-0653316; EC 269-915-2; EN300-248170; W-108655; Q24736495
|
|
| CAS | 112-18-5 | |
| PubChem CID | 8168 | |
| ChEMBL ID | CHEMBL109737 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 213.4 | ALogp: | 5.9 |
| HBD: | 0 | HBA: | 1 |
| Rotatable Bonds: | 11 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 3.2 | Aromatic Rings: | 0 |
| Heavy Atoms: | 15 | QED Weighted: | 0.438 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.61 | MDCK Permeability: | 0.00000971 |
| Pgp-inhibitor: | 0.015 | Pgp-substrate: | 0.006 |
| Human Intestinal Absorption (HIA): | 0.004 | 20% Bioavailability (F20%): | 0.32 |
| 30% Bioavailability (F30%): | 0.978 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.941 | Plasma Protein Binding (PPB): | 77.18% |
| Volume Distribution (VD): | 1.71 | Fu: | 20.44% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.341 | CYP1A2-substrate: | 0.895 |
| CYP2C19-inhibitor: | 0.105 | CYP2C19-substrate: | 0.982 |
| CYP2C9-inhibitor: | 0.02 | CYP2C9-substrate: | 0.584 |
| CYP2D6-inhibitor: | 0.927 | CYP2D6-substrate: | 0.914 |
| CYP3A4-inhibitor: | 0.041 | CYP3A4-substrate: | 0.278 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 6.956 | Half-life (T1/2): | 0.113 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.254 | Human Hepatotoxicity (H-HT): | 0.079 |
| Drug-inuced Liver Injury (DILI): | 0.17 | AMES Toxicity: | 0.007 |
| Rat Oral Acute Toxicity: | 0.835 | Maximum Recommended Daily Dose: | 0.033 |
| Skin Sensitization: | 0.944 | Carcinogencity: | 0.079 |
| Eye Corrosion: | 0.998 | Eye Irritation: | 0.805 |
| Respiratory Toxicity: | 0.974 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000281 | 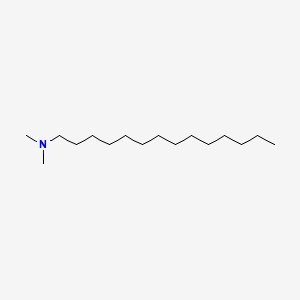 |
0.875 | D05ATI |  |
0.561 | ||
| ENC000421 | 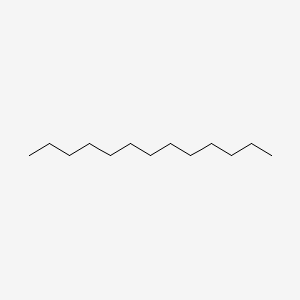 |
0.717 | D0Z5SM |  |
0.500 | ||
| ENC000327 |  |
0.681 | D07ILQ |  |
0.457 | ||
| ENC000276 | 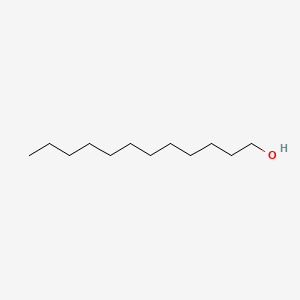 |
0.681 | D05QNO | 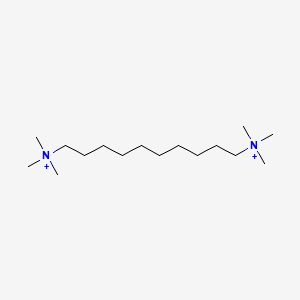 |
0.429 | ||
| ENC000422 |  |
0.673 | D00AOJ |  |
0.416 | ||
| ENC000272 |  |
0.652 | D00FGR |  |
0.410 | ||
| ENC000475 |  |
0.640 | D0O1PH |  |
0.403 | ||
| ENC001240 |  |
0.640 | D0Y8DP |  |
0.361 | ||
| ENC000423 |  |
0.635 | D0XN8C | 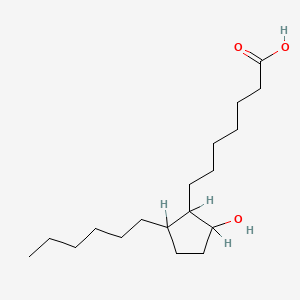 |
0.347 | ||
| ENC000274 | 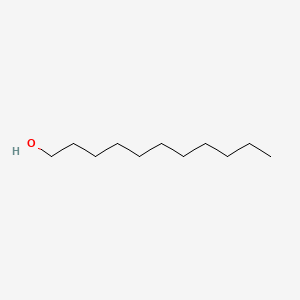 |
0.617 | D0MM8N | 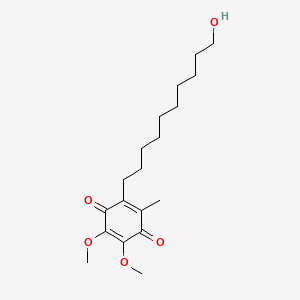 |
0.333 | ||