NPs Basic Information
|
Name |
Pentadecane
|
| Molecular Formula | C15H32 | |
| IUPAC Name* |
pentadecane
|
|
| SMILES |
CCCCCCCCCCCCCCC
|
|
| InChI |
InChI=1S/C15H32/c1-3-5-7-9-11-13-15-14-12-10-8-6-4-2/h3-15H2,1-2H3
|
|
| InChIKey |
YCOZIPAWZNQLMR-UHFFFAOYSA-N
|
|
| Synonyms |
Pentadecane; N-PENTADECANE; 629-62-9; Pentadekan; Pentadecane, n-; CHEBI:28897; 16H6K2S8M2; CH3-[CH2]13-CH3; NSC-172781; n-Pentadecane 100 microg/mL in Acetonitrile; Pentadecane, analytical standard; HSDB 5729; EINECS 211-098-1; MFCD00008990; NSC 172781; BRN 1698194; pentadecan; dipentylfumarate; UNII-16H6K2S8M2; Medicinal Plant; 1-Penfadecane,(S); Pentadecane, >=99%; DSSTox_CID_7268; PENTADECANE [INCI]; EC 211-098-1; ghl.PD_Mitscher_leg0.43; DSSTox_RID_78379; DSSTox_GSID_27268; N-PENTADECANE [HSDB]; 4-01-00-00529 (Beilstein Handbook Reference); CH3(CH2)13CH3; CHEMBL1234557; DTXSID6027268; Pentadecane_Ramanathan &Gurudeeban; Pentadecane, >=98.0% (GC); ZINC1531089; Tox21_300535; LMFA11000006; NSC172781; STL280516; AKOS015902386; NCGC00164185-01; NCGC00164185-02; NCGC00254392-01; CAS-629-62-9; LS-14458; FT-0700536; P0606; C08388; D97801; Q150831; 896D4B7E-BF33-4D54-82CE-7360D88E8DC8
|
|
| CAS | 629-62-9 | |
| PubChem CID | 12391 | |
| ChEMBL ID | CHEMBL1234557 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 212.41 | ALogp: | 7.7 |
| HBD: | 0 | HBA: | 0 |
| Rotatable Bonds: | 12 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 0.0 | Aromatic Rings: | 0 |
| Heavy Atoms: | 15 | QED Weighted: | 0.341 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.724 | MDCK Permeability: | 0.00000950 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.002 | 20% Bioavailability (F20%): | 0.403 |
| 30% Bioavailability (F30%): | 0.998 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.152 | Plasma Protein Binding (PPB): | 98.27% |
| Volume Distribution (VD): | 3.726 | Fu: | 1.65% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.413 | CYP1A2-substrate: | 0.199 |
| CYP2C19-inhibitor: | 0.436 | CYP2C19-substrate: | 0.096 |
| CYP2C9-inhibitor: | 0.127 | CYP2C9-substrate: | 0.943 |
| CYP2D6-inhibitor: | 0.212 | CYP2D6-substrate: | 0.065 |
| CYP3A4-inhibitor: | 0.195 | CYP3A4-substrate: | 0.053 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 4.637 | Half-life (T1/2): | 0.082 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.215 | Human Hepatotoxicity (H-HT): | 0.008 |
| Drug-inuced Liver Injury (DILI): | 0.222 | AMES Toxicity: | 0.007 |
| Rat Oral Acute Toxicity: | 0.038 | Maximum Recommended Daily Dose: | 0.029 |
| Skin Sensitization: | 0.947 | Carcinogencity: | 0.038 |
| Eye Corrosion: | 0.994 | Eye Irritation: | 0.943 |
| Respiratory Toxicity: | 0.531 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000379 |  |
0.935 | D0Z5SM |  |
0.644 | ||
| ENC000422 |  |
0.930 | D07ILQ |  |
0.585 | ||
| ENC000427 |  |
0.878 | D05ATI |  |
0.552 | ||
| ENC000421 |  |
0.860 | D00AOJ |  |
0.528 | ||
| ENC000400 |  |
0.827 | D00FGR |  |
0.494 | ||
| ENC000739 |  |
0.792 | D0O1PH |  |
0.453 | ||
| ENC000272 |  |
0.791 | D0T9TJ |  |
0.410 | ||
| ENC000428 |  |
0.782 | D05QNO |  |
0.400 | ||
| ENC000281 |  |
0.750 | D0P1RL |  |
0.390 | ||
| ENC000425 |  |
0.745 | D00MLW | 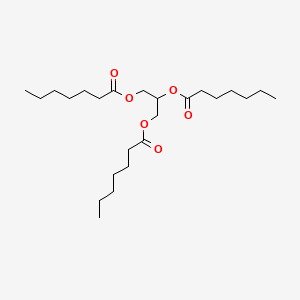 |
0.380 | ||