NPs Basic Information

|
Name |
3,4-Dihydro-2H-pyran
|
| Molecular Formula | C5H8O | |
| IUPAC Name* |
3,4-dihydro-2H-pyran
|
|
| SMILES |
C1CC=COC1
|
|
| InChI |
InChI=1S/C5H8O/c1-2-4-6-5-3-1/h2,4H,1,3,5H2
|
|
| InChIKey |
BUDQDWGNQVEFAC-UHFFFAOYSA-N
|
|
| Synonyms |
3,4-Dihydro-2H-pyran; 110-87-2; DIHYDROPYRAN; 3,4-Dihydropyran; 2H-3,4-Dihydropyran; 2,3-Dihydro-4H-pyran; Dihydro-2H-pyran; 2,3-Dihydropyran; 2H-Pyran, 3,4-dihydro-; 5,6-Dihydro-4H-pyran; 2H-Pyran, dihydro-; dihydropyrane; .delta.2-Dihydropyran; Pyran, dihydro-; 3,4-Dihydro-2-pyran; NSC 57860; T6V9N71IHX; 1,2-Pyran, 3,4-dihydro-; NSC-57860; NSC-73472; WLN: T6O BUTJ; 2,3-dihydropyrane; delta2-Dihydropyran; Dihydropyran (VAN); 3,4-dihydro-2H-pyrane; MFCD00006558; UNII-T6V9N71IHX; dihyrdopyran; dihyropyran; Dihydro-2h-pyran, 3,4-; 2,3dihydropyran; 2,3-dihyropyran; 2,4-dihydropyran; 4,5-dihydropyran; 3,4-dihydropyrane; EINECS 203-810-4; 2,3-dihydro-pyran; 3,4-dihydro-pyran; UN2376; 3,4dihydro-2H-pyran; 3,4-dihyro-2H-pyran; Pyran, 2,3-dihydro-; 3,4-dihydro-2h pyran; AI3-16497; 3, 4dihydro-2H-pyran; 3,4 dihydro-2H-pyran; 3,4-dihydro 2H-pyran; 3,4-dihydro-1H-pyran; 3.4-dihydro-2H-pyran; 2,3-dihydro-4H-pyrane; 3,4-Dihdro-2H-pyrane; 3,4-Dihydro(2H)pyran; DIHYDROPYRAN [MI]; 2,3-dihydro-4-H-pyran; 3,4- dihydro-2H-pyran; 3,4-dihydro-2 H-pyran; 3,4-dihydro-2-H-pyran; 3,4-dihydro-2H -pyran; 3,4-dihydro-2-H-pyrane; EC 203-810-4; 3,4-dihydro-[2H]-pyran; DSSTox_CID_21426; DSSTox_RID_79730; .delta.(Sup2)-Dihydropyran; 3,4 - Dihydro-2H-pyran; DSSTox_GSID_41426; CHEMBL3184439; DTXSID6041426; 3,4-Dihydro-2H-pyran, 97%; 2-PYRAN, 3,4-DIHYDRO-; ACT05913; AMY39440; NSC57860; NSC73472; STR01188; ZINC4726938; 2,3-DIHYDRO-.GAMMA.-PYRAN; Tox21_301188; STL146593; AKOS000121126; CS-W013755; NCGC00248323-01; NCGC00255086-01; BP-21473; CAS-110-87-2; DB-002492; D0555; EN300-26571; D78128; 2,3-Dihydropyran [UN2376] [Flammable liquid]; 3,4-Dihydro-2H-pyran, purum, >=95.0% (GC); Q419349; J-511179; F0001-0228
|
|
| CAS | 110-87-2; 3174- | |
| PubChem CID | 8080 | |
| ChEMBL ID | CHEMBL3184439 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 84.12 | ALogp: | 0.7 |
| HBD: | 0 | HBA: | 1 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 9.2 | Aromatic Rings: | 1 |
| Heavy Atoms: | 6 | QED Weighted: | 0.435 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.409 | MDCK Permeability: | 0.00002720 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.034 |
| Human Intestinal Absorption (HIA): | 0.003 | 20% Bioavailability (F20%): | 0.002 |
| 30% Bioavailability (F30%): | 0.248 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.996 | Plasma Protein Binding (PPB): | 26.80% |
| Volume Distribution (VD): | 1.98 | Fu: | 69.53% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.854 | CYP1A2-substrate: | 0.515 |
| CYP2C19-inhibitor: | 0.207 | CYP2C19-substrate: | 0.818 |
| CYP2C9-inhibitor: | 0.027 | CYP2C9-substrate: | 0.338 |
| CYP2D6-inhibitor: | 0.105 | CYP2D6-substrate: | 0.586 |
| CYP3A4-inhibitor: | 0.041 | CYP3A4-substrate: | 0.312 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 7.357 | Half-life (T1/2): | 0.733 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.024 | Human Hepatotoxicity (H-HT): | 0.297 |
| Drug-inuced Liver Injury (DILI): | 0.036 | AMES Toxicity: | 0.075 |
| Rat Oral Acute Toxicity: | 0.032 | Maximum Recommended Daily Dose: | 0.255 |
| Skin Sensitization: | 0.945 | Carcinogencity: | 0.958 |
| Eye Corrosion: | 0.786 | Eye Irritation: | 0.989 |
| Respiratory Toxicity: | 0.484 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC005597 | 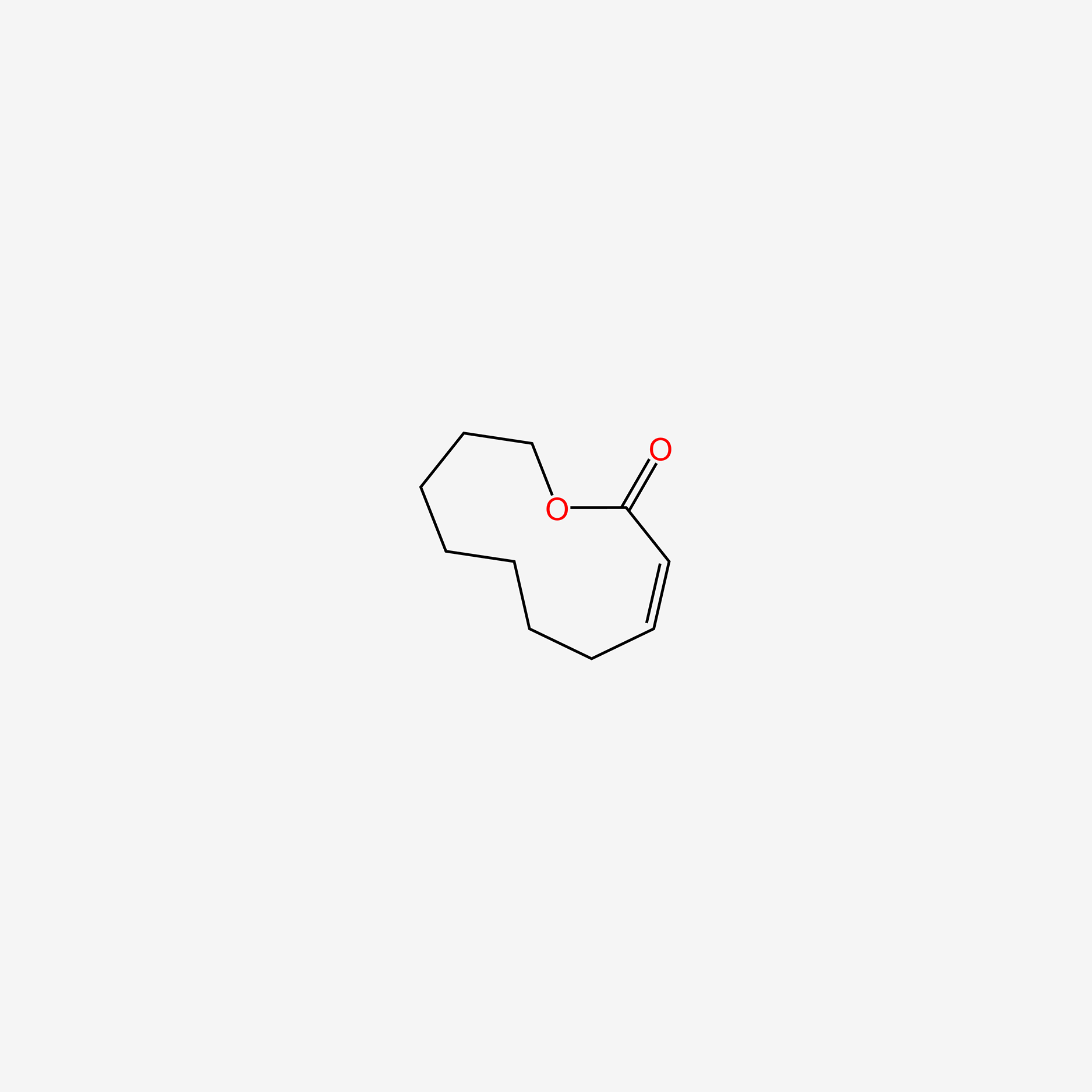 |
0.262 | D0Z8AA | 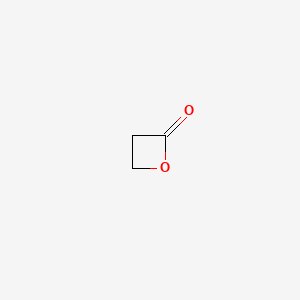 |
0.154 | ||
| ENC001730 |  |
0.257 | D07TGY | 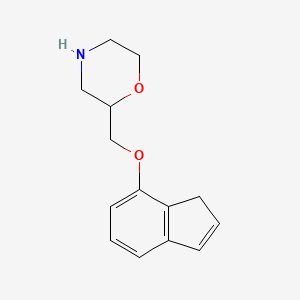 |
0.131 | ||
| ENC000540 |  |
0.243 | D05QIM | 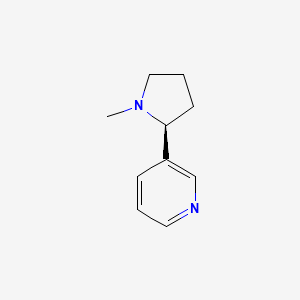 |
0.128 | ||
| ENC000184 | 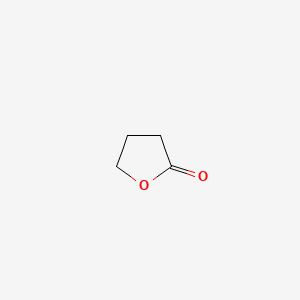 |
0.207 | D0M2MC |  |
0.120 | ||
| ENC001633 |  |
0.200 | D0CT9C |  |
0.118 | ||
| ENC002843 |  |
0.185 | D02TLO |  |
0.118 | ||
| ENC000753 | 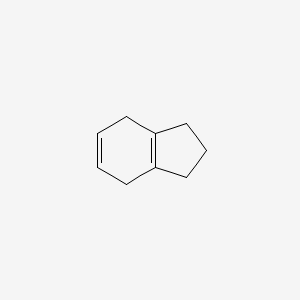 |
0.184 | D0T6SU |  |
0.115 | ||
| ENC004081 |  |
0.176 | D0V9JR | 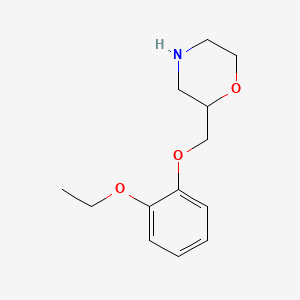 |
0.115 | ||
| ENC004080 |  |
0.176 | D0Y8PT | 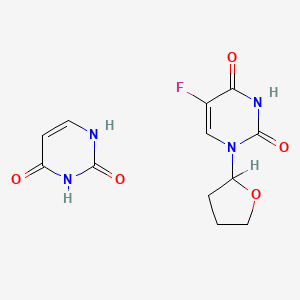 |
0.113 | ||
| ENC005373 |  |
0.174 | D07GRH | 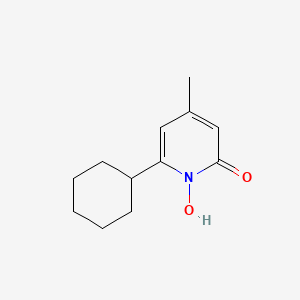 |
0.111 | ||