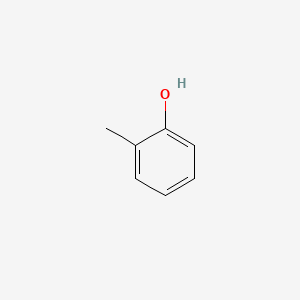
NPs Basic Information
|
Name |
o-Cresol
|
| Molecular Formula | C7H8O | |
| IUPAC Name* |
2-methylphenol
|
|
| SMILES |
CC1=CC=CC=C1O
|
|
| InChI |
InChI=1S/C7H8O/c1-6-4-2-3-5-7(6)8/h2-5,8H,1H3
|
|
| InChIKey |
QWVGKYWNOKOFNN-UHFFFAOYSA-N
|
|
| Synonyms |
o-cresol; 2-Methylphenol; 95-48-7; Orthocresol; 2-hydroxytoluene; 2-Cresol; Phenol, 2-methyl-; o-methylphenol; o-Cresylic acid; o-Oxytoluene; o-Toluol; 1-Hydroxy-2-methylbenzene; ortho-cresol; o-Hydroxytoluene; o-Methylphenylol; o-Kresol; Cresol, ortho-; Cresol, o-; 2-Hydroxy-1-methylbenzene; 2-methyl phenol; o-Kresol [German]; 2-methyl-phenol; Cresol, o-isomer; FEMA No. 3480; 1-Methyl-2-hydroxybenzene; NSC 23076; YW84DH5I7U; CHEBI:28054; MFCD00002226; NSC-23076; NSC-36809; TOLUENE,2-HYDROXY (ORTHO-CRESOL); o-Cresol [UN2076] [Poison, Corrosive]; DSSTox_CID_1808; DSSTox_RID_76341; WLN: QR B1; DSSTox_GSID_21808; hydroxy toluene; CAS-95-48-7; Orthocresol [NF]; CCRIS 646; HSDB 1813; EINECS 202-423-8; UNII-YW84DH5I7U; ortho cresol; Methyl phenol; 2-methyiphenol; AI3-00137; JZ0; O-Cresol,(S); Carvacrol derivative, 9; O-CRESOL [FHFI]; O-CRESOL [INCI]; o-Cresol, >=99%; O-CRESOL [MI]; ORTHOCRESOL [HSDB]; bmse000433; EC 202-423-8; 2-Methylphenol (o-cresol); ortho-cresol,2-methylphenol; SCHEMBL16002; MLS002454426; o-Cresol, analytical standard; BIDD:ER0677; CHEMBL46931; DTXSID8021808; BDBM248166; HMS2268O24; ORTHOCRESOL [USP IMPURITY]; ZINC901022; o-Cresol, for synthesis, 99.3%; 2-Methylphenol, analytical standard; NSC23076; NSC36809; Tox21_202305; Tox21_300021; STL194295; o-Cresol, ReagentPlus(R), >=99%; AKOS000119021; NCGC00091534-01; NCGC00091534-02; NCGC00091534-03; NCGC00091534-04; NCGC00254140-01; NCGC00259854-01; o-Cresol, SAJ first grade, >=97.0%; SMR001252248; 2-Methylphenol 100 microg/mL in Methanol; METACRESOL IMPURITY B [EP IMPURITY]; FT-0656046; EN300-19429; C01542; 1-Hydroxyl 2-Methyl Benzene, 2-Hydroxyl Toluene; Q312708; J-006098; J-523819; F0001-2271; Z104473822
|
|
| CAS | 95-48-7 | |
| PubChem CID | 335 | |
| ChEMBL ID | CHEMBL46931 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 108.14 | ALogp: | 2.0 |
| HBD: | 1 | HBA: | 1 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 20.2 | Aromatic Rings: | 1 |
| Heavy Atoms: | 8 | QED Weighted: | 0.54 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.284 | MDCK Permeability: | 0.00003070 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.011 |
| Human Intestinal Absorption (HIA): | 0.004 | 20% Bioavailability (F20%): | 0.112 |
| 30% Bioavailability (F30%): | 0.027 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.432 | Plasma Protein Binding (PPB): | 75.61% |
| Volume Distribution (VD): | 1.814 | Fu: | 16.98% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.858 | CYP1A2-substrate: | 0.916 |
| CYP2C19-inhibitor: | 0.427 | CYP2C19-substrate: | 0.651 |
| CYP2C9-inhibitor: | 0.085 | CYP2C9-substrate: | 0.709 |
| CYP2D6-inhibitor: | 0.387 | CYP2D6-substrate: | 0.872 |
| CYP3A4-inhibitor: | 0.032 | CYP3A4-substrate: | 0.286 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 14.833 | Half-life (T1/2): | 0.891 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.008 | Human Hepatotoxicity (H-HT): | 0.023 |
| Drug-inuced Liver Injury (DILI): | 0.076 | AMES Toxicity: | 0.118 |
| Rat Oral Acute Toxicity: | 0.811 | Maximum Recommended Daily Dose: | 0.041 |
| Skin Sensitization: | 0.718 | Carcinogencity: | 0.611 |
| Eye Corrosion: | 0.984 | Eye Irritation: | 0.996 |
| Respiratory Toxicity: | 0.651 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000021 | 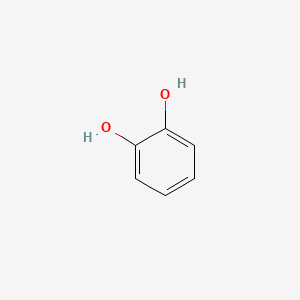 |
0.571 | D07HBX |  |
0.485 | ||
| ENC000179 | 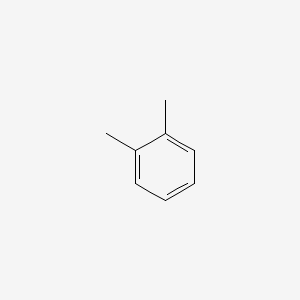 |
0.571 | D05OIS | 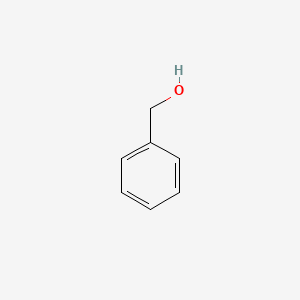 |
0.364 | ||
| ENC000033 | 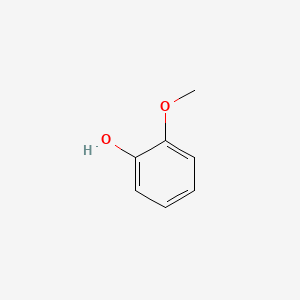 |
0.567 | D0T3NY | 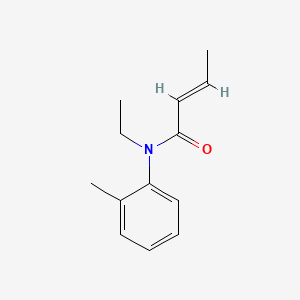 |
0.340 | ||
| ENC000408 |  |
0.516 | D0F5ZM | 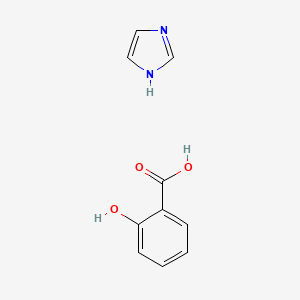 |
0.333 | ||
| ENC000166 |  |
0.516 | D0GY5Z | 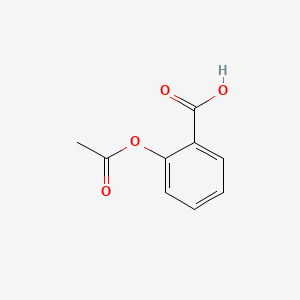 |
0.326 | ||
| ENC000407 | 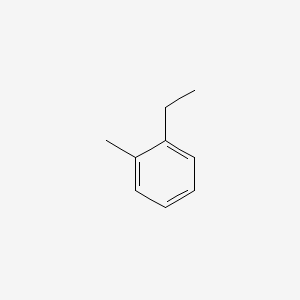 |
0.516 | D06LYG | 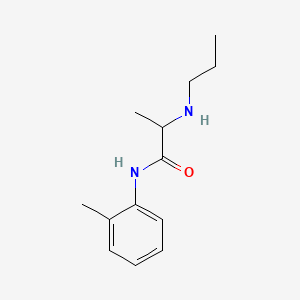 |
0.320 | ||
| ENC000104 |  |
0.486 | D03GET | 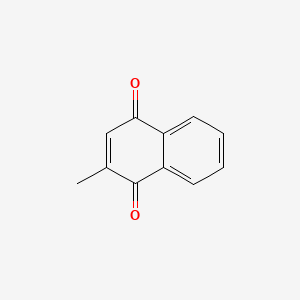 |
0.318 | ||
| ENC000365 |  |
0.485 | D0T3LF | 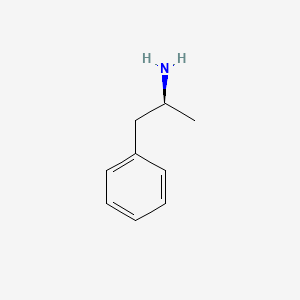 |
0.316 | ||
| ENC000108 | 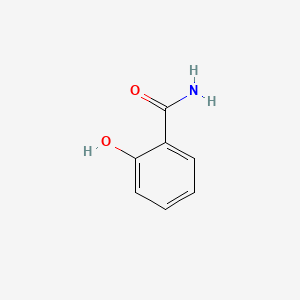 |
0.485 | D05BMG | 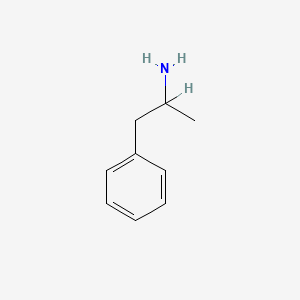 |
0.316 | ||
| ENC000917 | 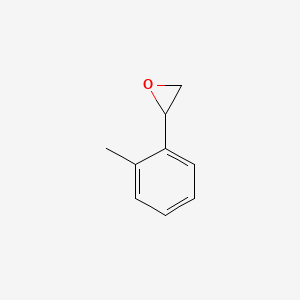 |
0.471 | D06DLI | 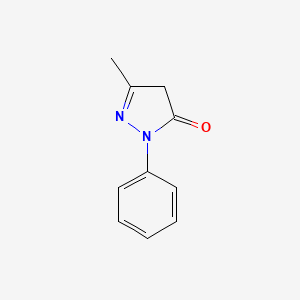 |
0.311 | ||