NPs Basic Information
|
Name |
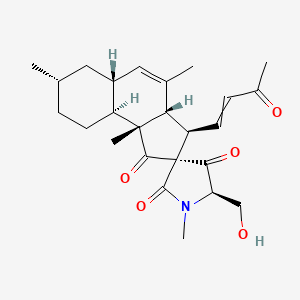
Pyrenosetin C
|
| Molecular Formula | C25H33NO5 | |
| IUPAC Name* |
(2R,3R,3aS,5'R,5aR,7S,9aS,9bR)-5'-(hydroxymethyl)-1',4,7,9b-tetramethyl-3-(3-oxobut-1-enyl)spiro[3,3a,5a,6,7,8,9,9a-octahydrocyclopenta[a]naphthalene-2,3'-pyrrolidine]-1,2',4'-trione
|
|
| SMILES |
C[C@H]1CC[C@H]2[C@H](C1)C=C([C@H]3[C@@]2(C(=O)[C@]4([C@@H]3C=CC(=O)C)C(=O)[C@H](N(C4=O)C)CO)C)C
|
|
| InChI |
InChI=1S/C25H33NO5/c1-13-6-8-17-16(10-13)11-14(2)20-18(9-7-15(3)28)25(22(30)24(17,20)4)21(29)19(12-27)26(5)23(25)31/h7,9,11,13,16-20,27H,6,8,10,12H2,1-5H3/t13-,16+,17-,18+,19+,20+,24+,25-/m0/s1
|
|
| InChIKey |
LQXNTMMEAPEMLM-DMLHXFJKSA-N
|
|
| Synonyms |
Pyrenosetin C
|
|
| CAS | NA | |
| PubChem CID | 156581893 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 427.5 | ALogp: | 2.7 |
| HBD: | 1 | HBA: | 5 |
| Rotatable Bonds: | 3 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 91.8 | Aromatic Rings: | 4 |
| Heavy Atoms: | 31 | QED Weighted: | 0.424 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.894 | MDCK Permeability: | 0.00002070 |
| Pgp-inhibitor: | 0.993 | Pgp-substrate: | 0.87 |
| Human Intestinal Absorption (HIA): | 0.294 | 20% Bioavailability (F20%): | 0.007 |
| 30% Bioavailability (F30%): | 0.04 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.931 | Plasma Protein Binding (PPB): | 69.31% |
| Volume Distribution (VD): | 0.881 | Fu: | 28.05% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.007 | CYP1A2-substrate: | 0.331 |
| CYP2C19-inhibitor: | 0.061 | CYP2C19-substrate: | 0.865 |
| CYP2C9-inhibitor: | 0.019 | CYP2C9-substrate: | 0.068 |
| CYP2D6-inhibitor: | 0.002 | CYP2D6-substrate: | 0.058 |
| CYP3A4-inhibitor: | 0.389 | CYP3A4-substrate: | 0.918 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 9.765 | Half-life (T1/2): | 0.4 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.016 | Human Hepatotoxicity (H-HT): | 0.07 |
| Drug-inuced Liver Injury (DILI): | 0.477 | AMES Toxicity: | 0.026 |
| Rat Oral Acute Toxicity: | 0.356 | Maximum Recommended Daily Dose: | 0.876 |
| Skin Sensitization: | 0.73 | Carcinogencity: | 0.927 |
| Eye Corrosion: | 0.024 | Eye Irritation: | 0.011 |
| Respiratory Toxicity: | 0.972 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC005775 | 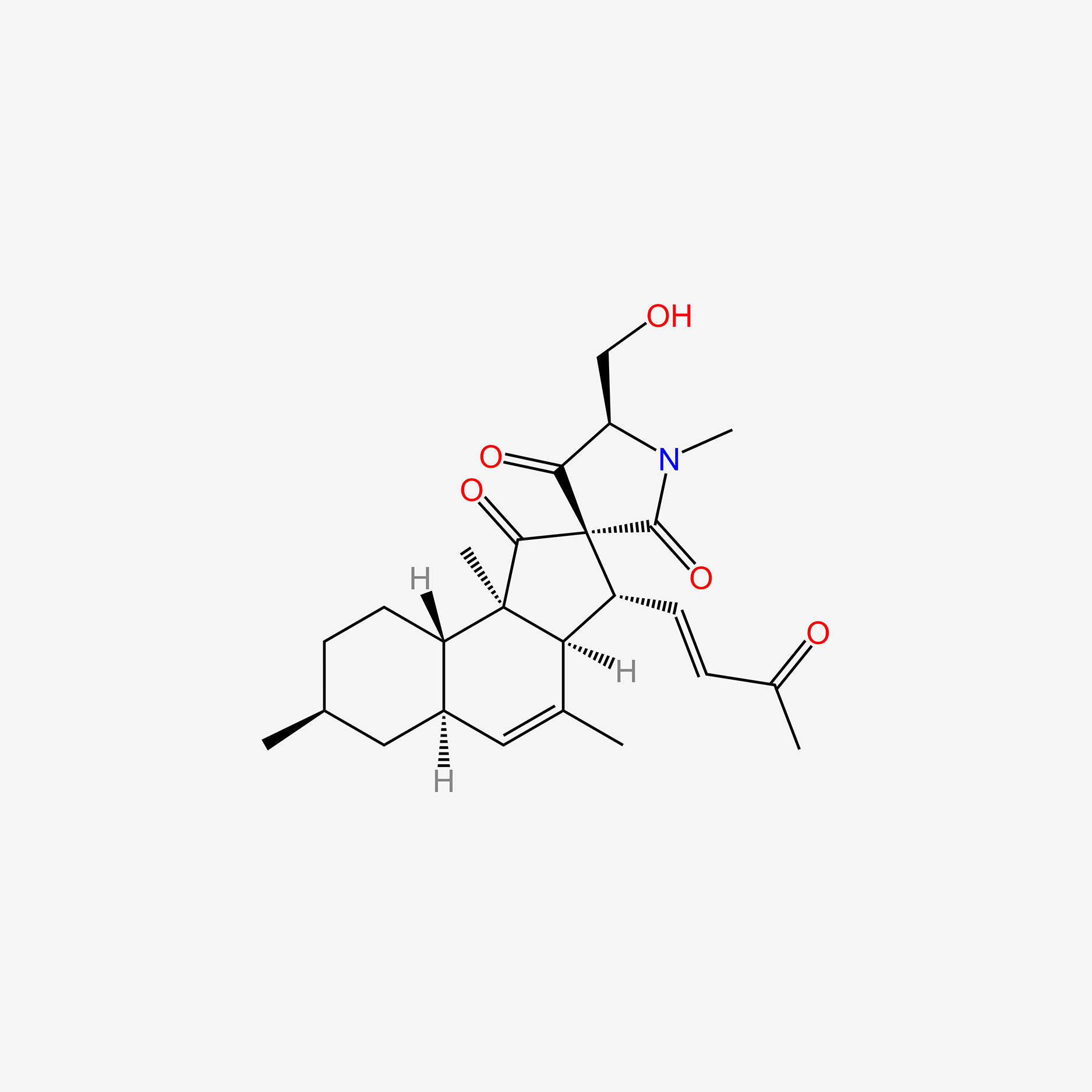 |
1.000 | D0E9KA | 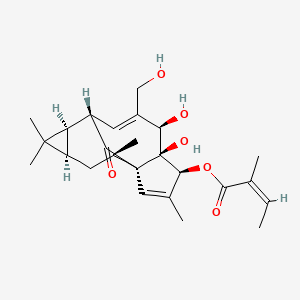 |
0.279 | ||
| ENC005774 | 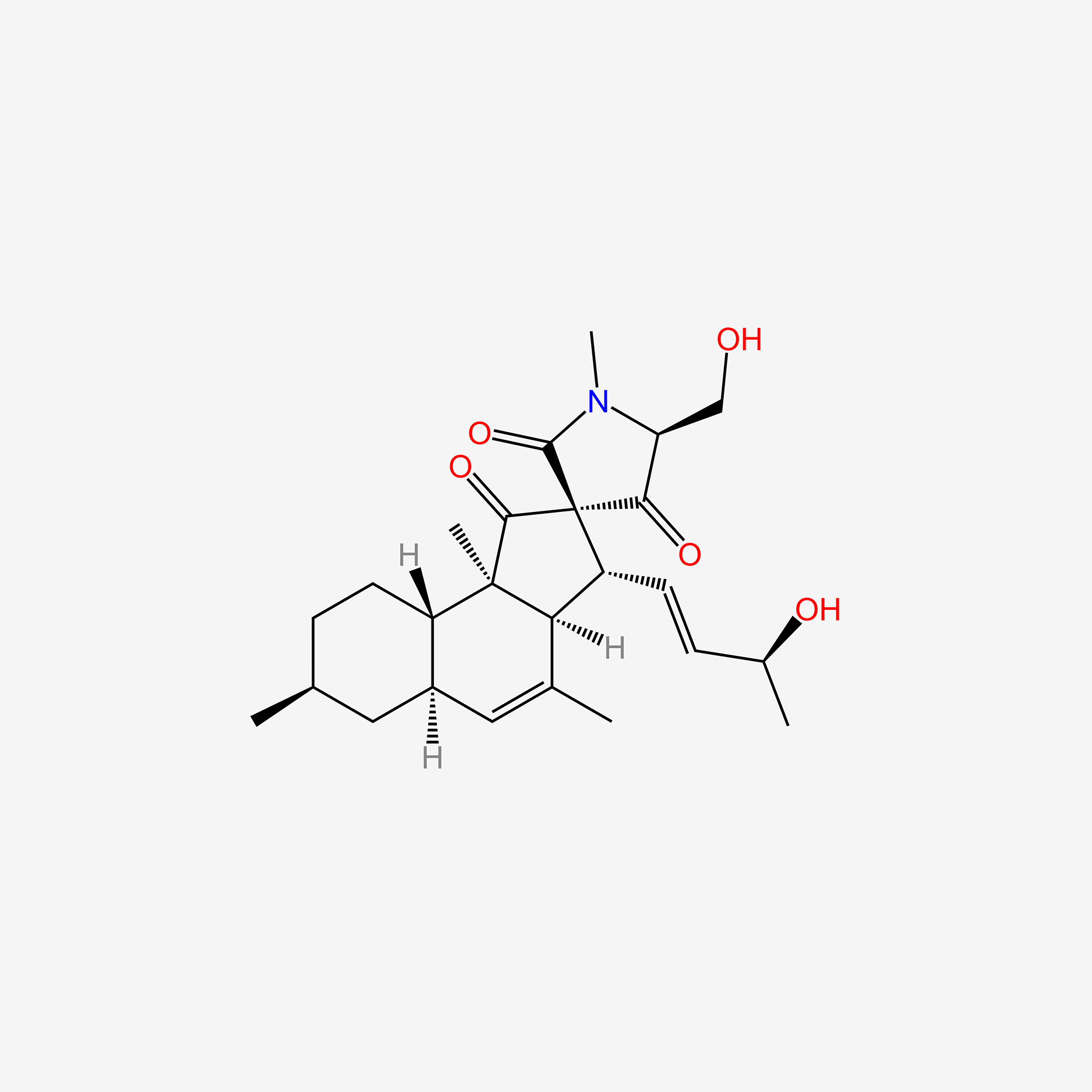 |
0.804 | D04SFH | 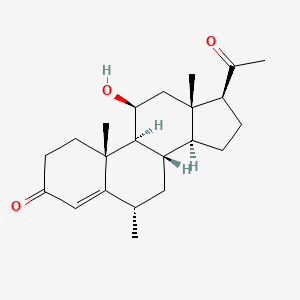 |
0.269 | ||
| ENC004321 | 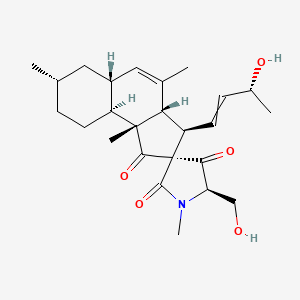 |
0.804 | D0I5DS | 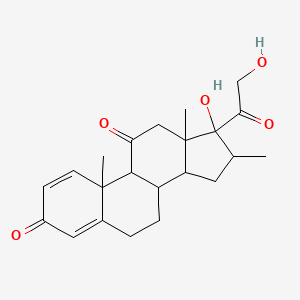 |
0.258 | ||
| ENC004320 | 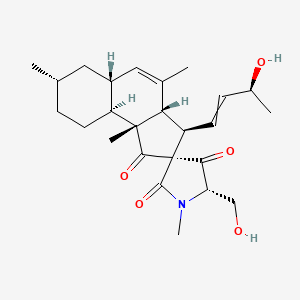 |
0.804 | D06AEO | 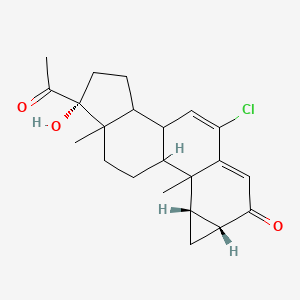 |
0.252 | ||
| ENC004339 | 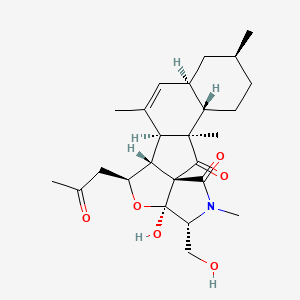 |
0.536 | D0W2EK | 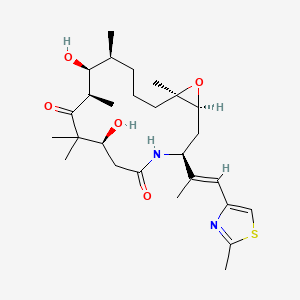 |
0.246 | ||
| ENC003818 | 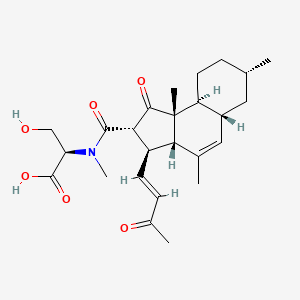 |
0.527 | D0D2TN |  |
0.238 | ||
| ENC005181 |  |
0.491 | D04GJN |  |
0.238 | ||
| ENC005182 |  |
0.491 | D0I2SD | 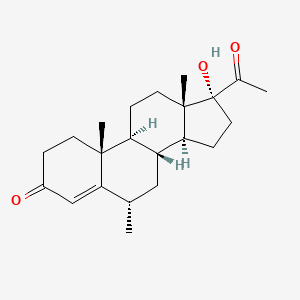 |
0.238 | ||
| ENC002818 | 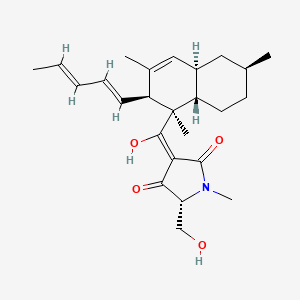 |
0.477 | D0X4RS | 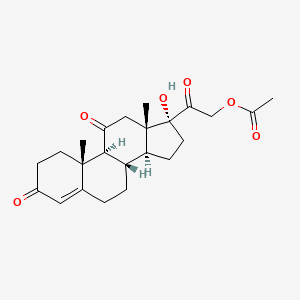 |
0.237 | ||
| ENC003817 | 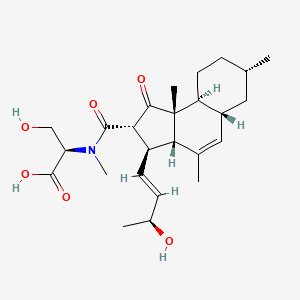 |
0.448 | D0V2JK |  |
0.236 | ||