NPs Basic Information
|
Name |
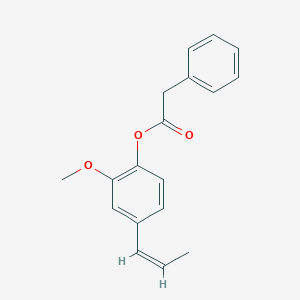
Isoeugenol phenylacetate
|
| Molecular Formula | C18H18O3 | |
| IUPAC Name* |
[2-methoxy-4-[(Z)-prop-1-enyl]phenyl] 2-phenylacetate
|
|
| SMILES |
C/C=C\C1=CC(=C(C=C1)OC(=O)CC2=CC=CC=C2)OC
|
|
| InChI |
InChI=1S/C18H18O3/c1-3-7-14-10-11-16(17(12-14)20-2)21-18(19)13-15-8-5-4-6-9-15/h3-12H,13H2,1-2H3/b7-3-
|
|
| InChIKey |
YYLCMLYMJHKLEJ-CLTKARDFSA-N
|
|
| Synonyms |
Isoeugenol phenylacetate; [2-methoxy-4-[(Z)-prop-1-enyl]phenyl] 2-phenylacetate; Isoeugenyl alpha-toluate; BENZENEACETIC ACID, 2-METHOXY-4-(1-PROPENYL)PHENYL ESTER; 4-Propenylguaiacyl phenylacetate; 2-Methoxy-4-propenylphenyl phenylacetate; 120-24-1; 2-Methoxy-4-prop-1-enylphenyl phenylacetate; 2-Methoxy-4-(1-propenyl)phenyl benzeneacetate; 2-Methoxy-4-(1-propen-1-yl)phenyl phenylacetate; z-isoeugenyl phenylacetate; SFI30QX6ZK; Acetic acid, phenyl-, 2-methoxy-4-propenylphenyl ester; (Z)-isoeugenyl phenylacetate; Benzeneacetic acid, 2-methoxy-4-(1-propen-1-yl)phenyl ester; FEMA 2477; CHEBI:174698; FEMA NO. 2477, Z-; ZINC14589773; ISOEUGENYL PHENYLACETATE, (Z)-; ISOEUGENYL PHENYL ACETATE, (Z)-; Q27289182; 2-methoxy-4-[(1Z)-prop-1-en-1-yl]phenyl 2-phenylacetate
|
|
| CAS | 120-24-1 | |
| PubChem CID | 20835508 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 282.3 | ALogp: | 4.0 |
| HBD: | 0 | HBA: | 3 |
| Rotatable Bonds: | 6 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 35.5 | Aromatic Rings: | 2 |
| Heavy Atoms: | 21 | QED Weighted: | 0.597 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.61 | MDCK Permeability: | 0.00002560 |
| Pgp-inhibitor: | 0.616 | Pgp-substrate: | 0.076 |
| Human Intestinal Absorption (HIA): | 0.005 | 20% Bioavailability (F20%): | 0.002 |
| 30% Bioavailability (F30%): | 0.694 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.645 | Plasma Protein Binding (PPB): | 97.25% |
| Volume Distribution (VD): | 0.411 | Fu: | 1.89% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.981 | CYP1A2-substrate: | 0.43 |
| CYP2C19-inhibitor: | 0.964 | CYP2C19-substrate: | 0.341 |
| CYP2C9-inhibitor: | 0.871 | CYP2C9-substrate: | 0.415 |
| CYP2D6-inhibitor: | 0.12 | CYP2D6-substrate: | 0.206 |
| CYP3A4-inhibitor: | 0.679 | CYP3A4-substrate: | 0.601 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 13.197 | Half-life (T1/2): | 0.814 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.016 | Human Hepatotoxicity (H-HT): | 0.157 |
| Drug-inuced Liver Injury (DILI): | 0.847 | AMES Toxicity: | 0.066 |
| Rat Oral Acute Toxicity: | 0.03 | Maximum Recommended Daily Dose: | 0.023 |
| Skin Sensitization: | 0.884 | Carcinogencity: | 0.45 |
| Eye Corrosion: | 0.011 | Eye Irritation: | 0.619 |
| Respiratory Toxicity: | 0.056 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC001446 | 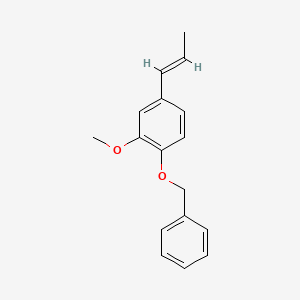 |
0.742 | D0E6OC | 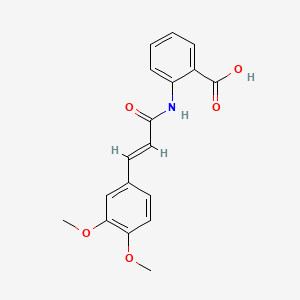 |
0.477 | ||
| ENC001461 | 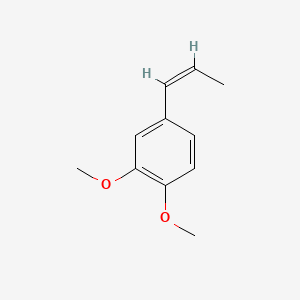 |
0.477 | D0Y7EM | 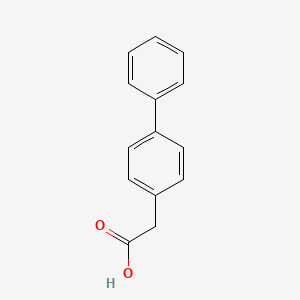 |
0.359 | ||
| ENC000208 | 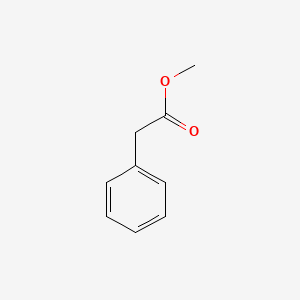 |
0.444 | D0G1VX |  |
0.354 | ||
| ENC001291 | 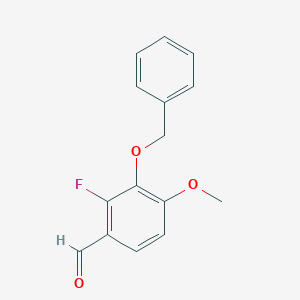 |
0.443 | D09VXM | 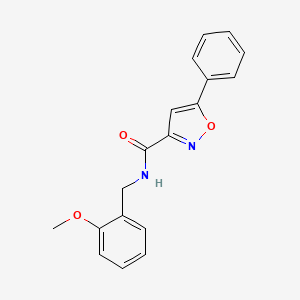 |
0.351 | ||
| ENC001523 | 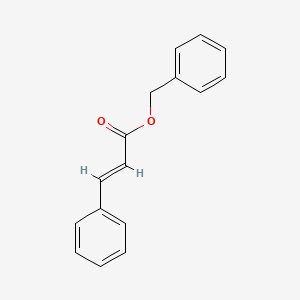 |
0.395 | D0L5PO | 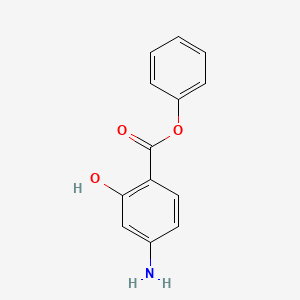 |
0.350 | ||
| ENC001456 | 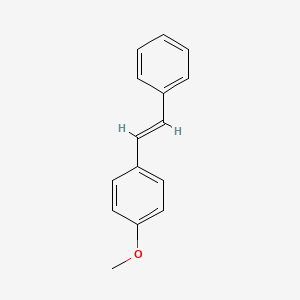 |
0.390 | D0S5RZ | 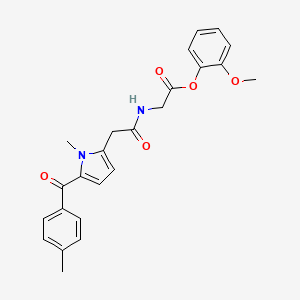 |
0.349 | ||
| ENC001737 |  |
0.381 | D08CCE |  |
0.348 | ||
| ENC000209 |  |
0.380 | D0KS6W |  |
0.337 | ||
| ENC000218 | 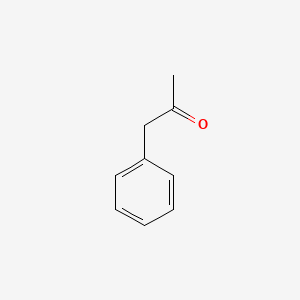 |
0.375 | D0IN7I | 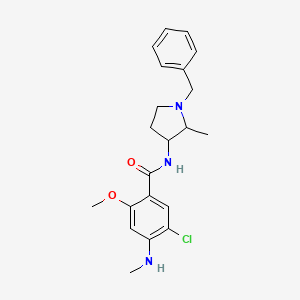 |
0.333 | ||
| ENC000823 | 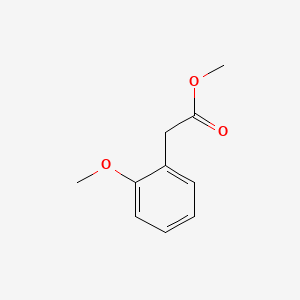 |
0.371 | D0H6TP | 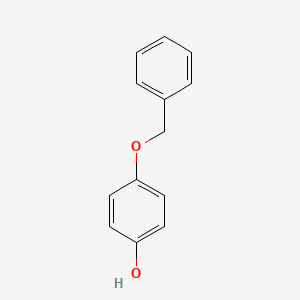 |
0.333 | ||