NPs Basic Information
|
Name |
Ginsenoside Rb2
|
| Molecular Formula | C53H90O22 | |
| IUPAC Name* |
(2S,3R,4S,5S,6R)-2-[(2R,3R,4S,5S,6R)-4,5-dihydroxy-6-(hydroxymethyl)-2-[[(3S,5R,8R,9R,10R,12R,13R,14R,17S)-12-hydroxy-4,4,8,10,14-pentamethyl-17-[(2S)-6-methyl-2-[(2S,3R,4S,5S,6R)-3,4,5-trihydroxy-6-[[(2S,3R,4S,5S)-3,4,5-trihydroxyoxan-2-yl]oxymethyl]oxan-2-yl]oxyhept-5-en-2-yl]-2,3,5,6,7,9,11,12,13,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-3-yl]oxy]oxan-3-yl]oxy-6-(hydroxymethyl)oxane-3,4,5-triol
|
|
| SMILES |
CC(=CCC[C@@](C)([C@H]1CC[C@@]2([C@@H]1[C@@H](C[C@H]3[C@]2(CC[C@@H]4[C@@]3(CC[C@@H](C4(C)C)O[C@H]5[C@@H]([C@H]([C@@H]([C@H](O5)CO)O)O)O[C@H]6[C@@H]([C@H]([C@@H]([C@H](O6)CO)O)O)O)C)C)O)C)O[C@H]7[C@@H]([C@H]([C@@H]([C@H](O7)CO[C@H]8[C@@H]([C@H]([C@H](CO8)O)O)O)O)O)O)C
|
|
| InChI |
InChI=1S/C53H90O22/c1-23(2)10-9-14-53(8,75-47-43(67)39(63)37(61)29(72-47)22-69-45-41(65)34(58)26(57)21-68-45)24-11-16-52(7)33(24)25(56)18-31-50(5)15-13-32(49(3,4)30(50)12-17-51(31,52)6)73-48-44(40(64)36(60)28(20-55)71-48)74-46-42(66)38(62)35(59)27(19-54)70-46/h10,24-48,54-67H,9,11-22H2,1-8H3/t24-,25+,26-,27+,28+,29+,30-,31+,32-,33-,34-,35+,36+,37+,38-,39-,40-,41+,42+,43+,44+,45-,46-,47-,48-,50-,51+,52+,53-/m0/s1
|
|
| InChIKey |
NODILNFGTFIURN-GZPRDHCNSA-N
|
|
| Synonyms |
Ginsenoside Rb2; 11021-13-9; Ginsenoside C; GinsenosideRb2; Ginsenoside-Rb2; NSC 308878; CHEBI:77152; (20S)-ginsenoside Rb2; N219O0L31C; UNII-N219O0L31C; NSC-308878; EINECS 234-251-4; MFCD00221755; CHEMBL449303; HY-N0040; BDBM50450908; Ginsenoside Rb2, analytical standard; s9079; AKOS037514668; CCG-270635; CS-3830; DB06748; NCGC00485981-01; AC-34662; AS-56619; 021G139; Q27891169; (3beta,12beta)-20-{[6-O-(alpha-L-arabinopyranosyl)-beta-D-glucopyranosyl]oxy}-12-hydroxydammar-24-en-3-yl 2-O-beta-D-glucopyranosyl-beta-D-glucopyranoside; .BETA.-D-GLUCOPYRANOSIDE, (3.BETA.,12.BETA.)-20-((6-O-.ALPHA.-L-ARABINOPYRANOSYL-.BETA.-D-GLUCOPYRANOSYL)OXY)-12-HYDROXYDAMMAR-24-EN-3-YL 2-O-.BETA.-D-GLUCOPYRANOSYL-; 20-((6-O-alpha-L-Arabinopyranosyl-beta-D-glucopyranosyl)oxy)-12beta-hydroxydammar-24-en-3beta-yl 2-O-beta-D-glucopyranosyl-beta-D-glucopyranoside; 20-[alpha-L-arabinopyranosyl-(1->2)-beta-D glucopyranosyloxy]-3beta-[beta-D-glucopyranosyl-(1->2)-beta-D glucopyranosyloxy]dammar-24-en-12beta-ol; beta-D-Glucopyranoside, (3-beta,12-beta)-20-((6-O-alpha-L-arabinopyranosyl-beta-D-glucopyranosyl)oxy)-12-hydroxydammar-24-en-3-yl 2-O-beta-D-glucopyranosyl-; GINSENOSIDE RB2 (CONSTITUENT OF AMERICAN GINSENG, ASIAN GINSENG, AND TIENCHI GINSENG) [DSC]
|
|
| CAS | 11021-13-9 | |
| PubChem CID | 6917976 | |
| ChEMBL ID | CHEMBL449303 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 1079.3 | ALogp: | 0.3 |
| HBD: | 14 | HBA: | 22 |
| Rotatable Bonds: | 15 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 357.0 | Aromatic Rings: | 8 |
| Heavy Atoms: | 75 | QED Weighted: | 0.069 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -6.046 | MDCK Permeability: | 0.00012679 |
| Pgp-inhibitor: | 0.998 | Pgp-substrate: | 0.05 |
| Human Intestinal Absorption (HIA): | 0.997 | 20% Bioavailability (F20%): | 0.842 |
| 30% Bioavailability (F30%): | 0.999 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.029 | Plasma Protein Binding (PPB): | 72.60% |
| Volume Distribution (VD): | -0.15 | Fu: | 11.26% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0 | CYP1A2-substrate: | 0.083 |
| CYP2C19-inhibitor: | 0 | CYP2C19-substrate: | 0.07 |
| CYP2C9-inhibitor: | 0 | CYP2C9-substrate: | 0.004 |
| CYP2D6-inhibitor: | 0.003 | CYP2D6-substrate: | 0.05 |
| CYP3A4-inhibitor: | 0.005 | CYP3A4-substrate: | 0.003 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 0.089 | Half-life (T1/2): | 0.714 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.337 | Human Hepatotoxicity (H-HT): | 0.089 |
| Drug-inuced Liver Injury (DILI): | 0.003 | AMES Toxicity: | 0.059 |
| Rat Oral Acute Toxicity: | 0.076 | Maximum Recommended Daily Dose: | 0.002 |
| Skin Sensitization: | 0.925 | Carcinogencity: | 0.006 |
| Eye Corrosion: | 0.003 | Eye Irritation: | 0.011 |
| Respiratory Toxicity: | 0.076 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
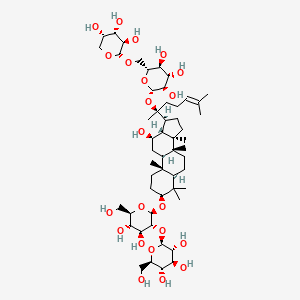
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC002245 | 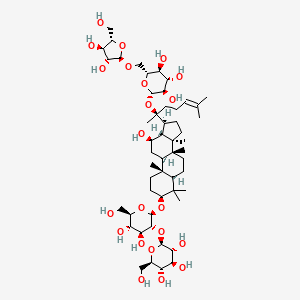 |
0.906 | D0P2IT | 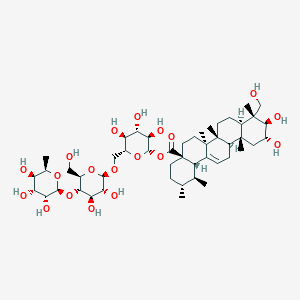 |
0.494 | ||
| ENC001933 | 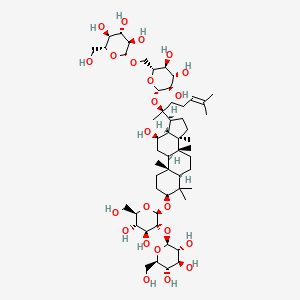 |
0.894 | D07QQD |  |
0.482 | ||
| ENC002655 |  |
0.859 | D04MRG |  |
0.465 | ||
| ENC002180 |  |
0.841 | D0A8RX |  |
0.459 | ||
| ENC001938 |  |
0.729 | D07ORO |  |
0.388 | ||
| ENC001939 |  |
0.688 | D07XBE |  |
0.375 | ||
| ENC000865 |  |
0.543 | D0P6IK |  |
0.340 | ||
| ENC001918 |  |
0.509 | D0AD5C |  |
0.338 | ||
| ENC002246 |  |
0.462 | D07BSE |  |
0.337 | ||
| ENC002265 |  |
0.437 | D07TGN |  |
0.336 | ||