NPs Basic Information
|
Name |
penitrem A
|
| Molecular Formula | C37H44ClNO6 | |
| IUPAC Name* |
(1S,2R,5S,6S,8R,9S,10R,12S,15R,16S,25R,27S,28R)-21-chloro-15,16,33,33-tetramethyl-24-methylidene-10-prop-1-en-2-yl-7,11,32-trioxa-18-azadecacyclo[25.4.2.02,16.05,15.06,8.06,12.017,31.019,30.022,29.025,28]tritriaconta-17(31),19,21,29-tetraene-5,9,28-triol
|
|
| SMILES |
CC(=C)[C@@H]1[C@@H]([C@@H]2[C@@]3(O2)[C@@H](O1)CC[C@]4([C@]3(CC[C@@H]5[C@@]4(C6=C7[C@H]5OC([C@H]8C[C@H]9[C@@]8(C1=C7C(=CC(=C1CC9=C)Cl)N6)O)(C)C)C)O)C)O
|
|
| InChI |
InChI=1S/C37H44ClNO6/c1-15(2)28-27(40)31-37(45-31)23(43-28)9-10-33(6)34(7)18(8-11-35(33,37)41)29-25-24-21(39-30(25)34)14-20(38)17-12-16(3)19-13-22(32(4,5)44-29)36(19,42)26(17)24/h14,18-19,22-23,27-29,31,39-42H,1,3,8-13H2,2,4-7H3/t18-,19+,22+,23-,27-,28+,29-,31+,33+,34+,35-,36+,37-/m0/s1
|
|
| InChIKey |
JDUWHZOLEDOQSR-JKPSMKLGSA-N
|
|
| Synonyms |
penitrem A; 12627-35-9; Tremortin A; 244AU85PR7; (1S,2R,5S,6S,8R,9S,10R,12S,15R,16S,25R,27S,28R)-21-chloro-15,16,33,33-tetramethyl-24-methylidene-10-prop-1-en-2-yl-7,11,32-trioxa-18-azadecacyclo[25.4.2.02,16.05,15.06,8.06,12.017,31.019,30.022,29.025,28]tritriaconta-17(31),19,21,29-tetraene-5,9,28-triol; NSC 354845; 2R,3S,3aR,4aS,4bS,6aR,7S,7dR,8R,9aR,14bS,14cR,16aS)-12-chloro-3,3a,6a,8,9,9a,10,11,14,14b,14c,15,16,16a-tetradecahydro-14b,14c,17,17-tetramethyl-10-methylene-2-(1-methylethenyl)-7,8-(epoxymethano)-2H,6H-cyclobuta[5,6]benz[1,2-e]oxireno[4',4'a]-1-benzopyrano[5',6':6,7]indeno[1,2-b]indole-3,4b,7d(5H,7H)-triol; C37H44ClNO6; NSC-354845; Penitrem A_120257; CBiol_001846; BSPBio_001431; KBioGR_000151; KBioSS_000151; SCHEMBL353317; UNII-244AU85PR7; CHEMBL2374089; KBio2_000151; KBio2_002719; KBio2_005287; KBio3_000301; KBio3_000302; DTXSID00320093; CHEBI:176948; Bio1_000132; Bio1_000621; Bio1_001110; Bio2_000151; Bio2_000631; HMS1361H13; HMS3402H13; HY-N6776; HB1057; MFCD12547934; ZINC67902768; AKOS024458256; IDI1_033901; QTL1_000065; NCGC00163403-01; NCGC00163403-02; NCGC00163403-03; Penitrem A, >=95% (HPLC and TLC); BP162742; CS-0029226; C20070; BRD-K03842655-001-02-1; Q63409025
|
|
| CAS | 12627-35-9 | |
| PubChem CID | 6610243 | |
| ChEMBL ID | CHEMBL2374089 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 634.2 | ALogp: | 3.8 |
| HBD: | 4 | HBA: | 6 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 108.0 | Aromatic Rings: | 10 |
| Heavy Atoms: | 45 | QED Weighted: | 0.233 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.383 | MDCK Permeability: | 0.00001060 |
| Pgp-inhibitor: | 0.979 | Pgp-substrate: | 0.995 |
| Human Intestinal Absorption (HIA): | 0.033 | 20% Bioavailability (F20%): | 0.866 |
| 30% Bioavailability (F30%): | 0.91 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.962 | Plasma Protein Binding (PPB): | 90.78% |
| Volume Distribution (VD): | 2.19 | Fu: | 3.24% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.029 | CYP1A2-substrate: | 0.973 |
| CYP2C19-inhibitor: | 0.084 | CYP2C19-substrate: | 0.638 |
| CYP2C9-inhibitor: | 0.264 | CYP2C9-substrate: | 0.062 |
| CYP2D6-inhibitor: | 0.036 | CYP2D6-substrate: | 0.417 |
| CYP3A4-inhibitor: | 0.629 | CYP3A4-substrate: | 0.883 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 5.996 | Half-life (T1/2): | 0.045 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.909 | Human Hepatotoxicity (H-HT): | 0.62 |
| Drug-inuced Liver Injury (DILI): | 0.556 | AMES Toxicity: | 0.022 |
| Rat Oral Acute Toxicity: | 0.985 | Maximum Recommended Daily Dose: | 0.992 |
| Skin Sensitization: | 0.224 | Carcinogencity: | 0.909 |
| Eye Corrosion: | 0.003 | Eye Irritation: | 0.01 |
| Respiratory Toxicity: | 0.989 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
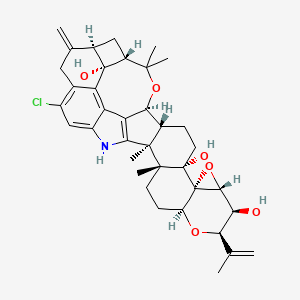
| ENC001508 | 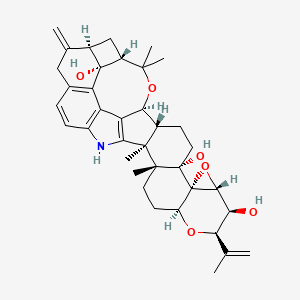 |
0.836 | D0KR9U |  |
0.224 | ||
| ENC001507 | 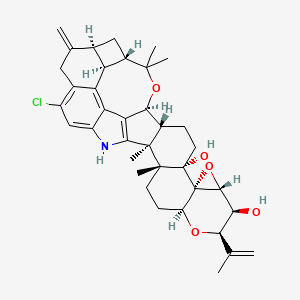 |
0.783 | D06IIB | 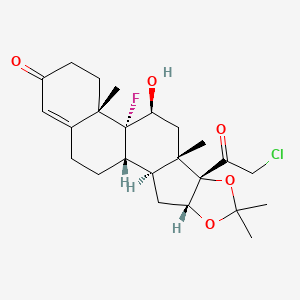 |
0.224 | ||
| ENC005404 | 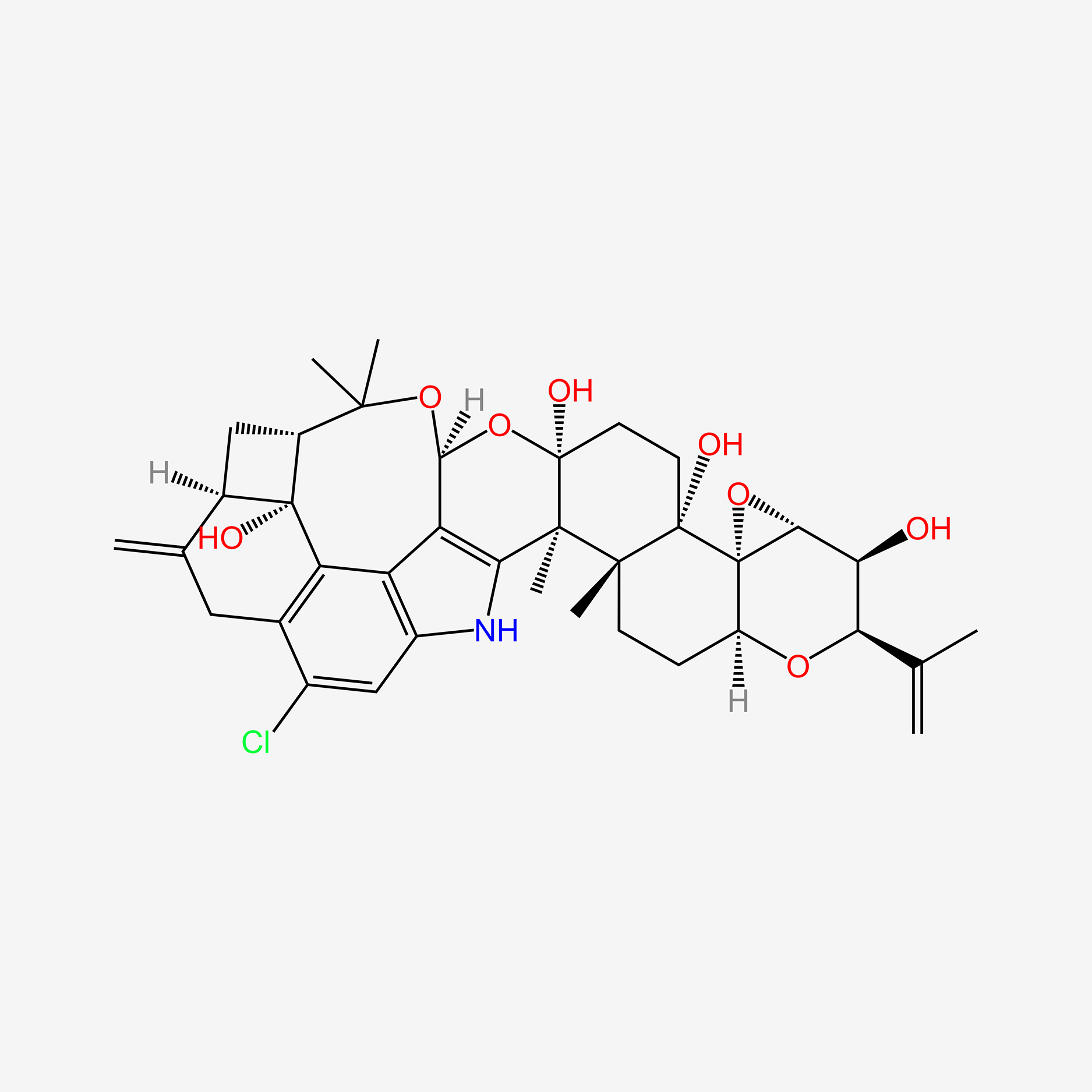 |
0.769 | D06AEO | 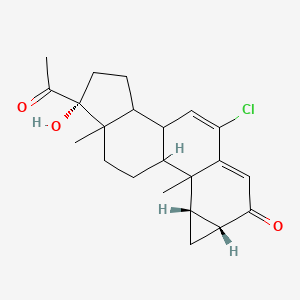 |
0.219 | ||
| ENC001499 | 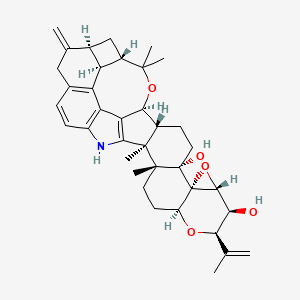 |
0.671 | D0W2EK |  |
0.218 | ||
| ENC003830 |  |
0.660 | D0AR3J |  |
0.216 | ||
| ENC003453 |  |
0.578 | D0Y2YP |  |
0.216 | ||
| ENC003831 |  |
0.546 | D0M2QH |  |
0.215 | ||
| ENC001486 |  |
0.497 | D0X7XG | 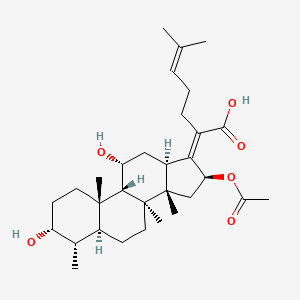 |
0.212 | ||
| ENC003833 | 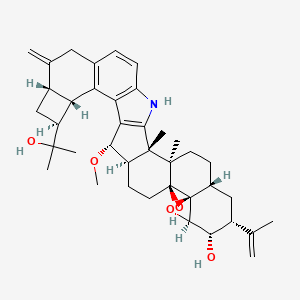 |
0.450 | D0S0NK | 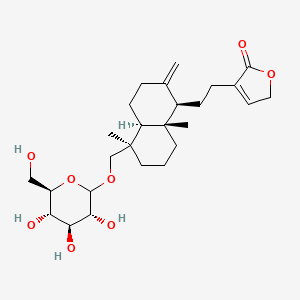 |
0.211 | ||
| ENC005557 | 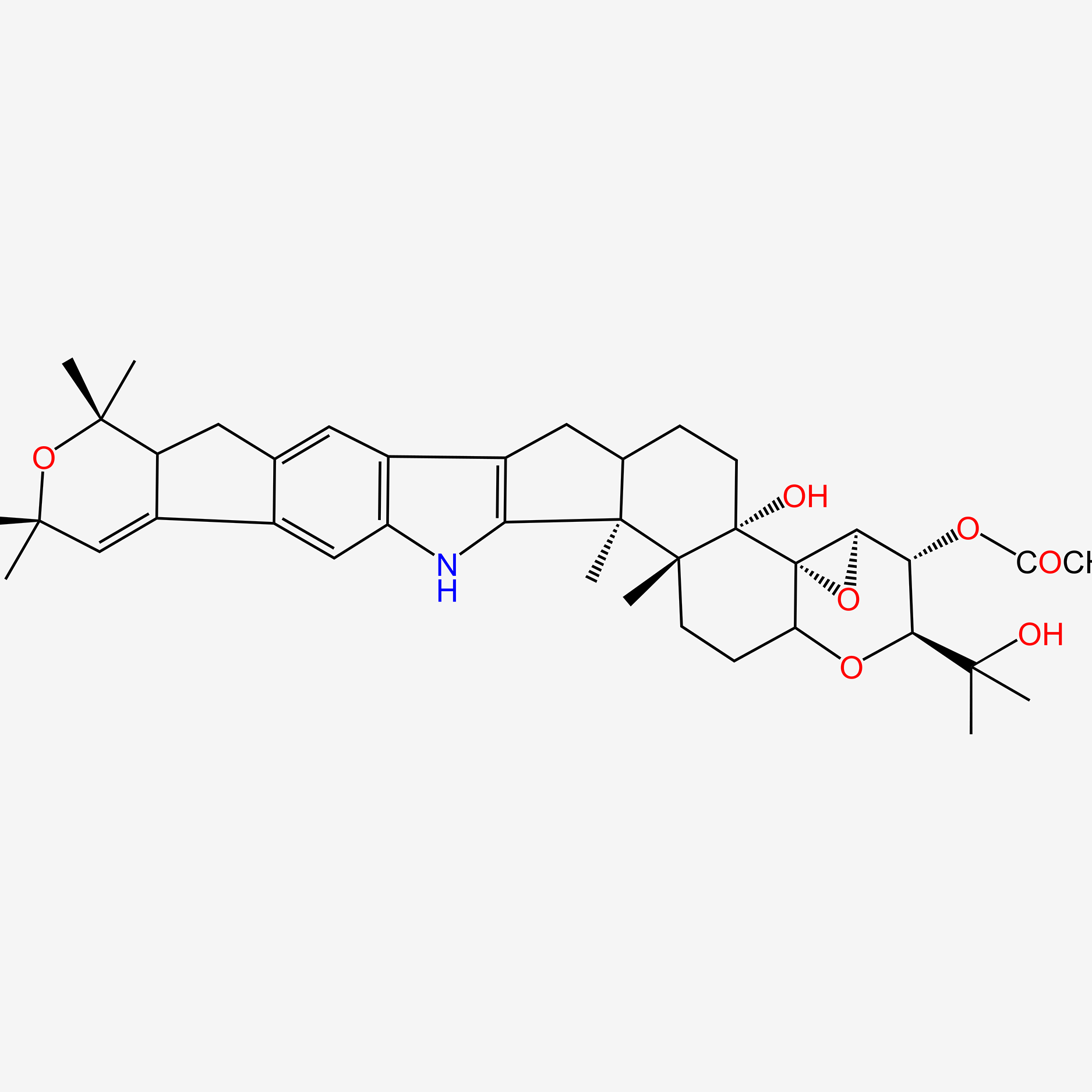 |
0.398 | D0I2SD |  |
0.208 | ||