NPs Basic Information
|
Name |
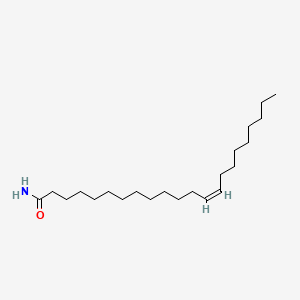
Erucamide
|
| Molecular Formula | C22H43NO | |
| IUPAC Name* |
(Z)-docos-13-enamide
|
|
| SMILES |
CCCCCCCC/C=C\CCCCCCCCCCCC(=O)N
|
|
| InChI |
InChI=1S/C22H43NO/c1-2-3-4-5-6-7-8-9-10-11-12-13-14-15-16-17-18-19-20-21-22(23)24/h9-10H,2-8,11-21H2,1H3,(H2,23,24)/b10-9-
|
|
| InChIKey |
UAUDZVJPLUQNMU-KTKRTIGZSA-N
|
|
| Synonyms |
ERUCAMIDE; (Z)-Docos-13-enamide; 112-84-5; Erucylamide; Erucyl amide; Erucic acid amide; cis-13-Docosenamide; cis-13-Docosenoamide; 13-Docosenamide, (13Z)-; 13-Docosenamide, (Z)-; erucilamide; (Z)-13-Docosenamide; Adogen 58; plastic additive 13; 13Z-Docosenamide; (13Z)-docos-13-enamide; (13Z)-docosenamide; (13Z)-13-Docosenamide; 0V89VY25BN; CHEMBL1232565; Unislip 1753; erucic amide; Erucoyl amide; Crodamide E; Kemamide E; Armid E; HSDB 5577; EINECS 204-009-2; UNII-0V89VY25BN; Euracamide; Petrac eramide; Plastic additive 21; Erucamide; erucylamide; 13-Docosenamide, cis-; ERUCAMIDE [HSDB]; ERUCAMIDE [INCI]; EC 204-009-2; SCHEMBL15126; Erucamide, analytical standard; (13Z)-13-Docosenamide #; ERUCAMIDE (ERUCYLAMIDE); DTXSID4026929; CHEBI:142245; HMS3649L13; BDBM50463965; LMFA08010028; MFCD00882379; ZINC14232715; AKOS016009791; PLASTIC ADDITIVE 13 [USP-RS]; AS-75453; D1008; A802673; SR-01000946673; SR-01000946673-1; W-108637; Q27231146; Plastic additive 13, United States Pharmacopeia (USP) Reference Standard
|
|
| CAS | 112-84-5 | |
| PubChem CID | 5365371 | |
| ChEMBL ID | CHEMBL1232565 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 337.6 | ALogp: | 8.8 |
| HBD: | 1 | HBA: | 1 |
| Rotatable Bonds: | 19 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 43.1 | Aromatic Rings: | 0 |
| Heavy Atoms: | 24 | QED Weighted: | 0.2 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.041 | MDCK Permeability: | 0.00002410 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0 |
| Human Intestinal Absorption (HIA): | 0.013 | 20% Bioavailability (F20%): | 0.938 |
| 30% Bioavailability (F30%): | 0.998 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.018 | Plasma Protein Binding (PPB): | 99.38% |
| Volume Distribution (VD): | 2.407 | Fu: | 0.73% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.122 | CYP1A2-substrate: | 0.19 |
| CYP2C19-inhibitor: | 0.278 | CYP2C19-substrate: | 0.051 |
| CYP2C9-inhibitor: | 0.101 | CYP2C9-substrate: | 0.944 |
| CYP2D6-inhibitor: | 0.342 | CYP2D6-substrate: | 0.277 |
| CYP3A4-inhibitor: | 0.35 | CYP3A4-substrate: | 0.029 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 4.099 | Half-life (T1/2): | 0.268 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.653 | Human Hepatotoxicity (H-HT): | 0.034 |
| Drug-inuced Liver Injury (DILI): | 0.034 | AMES Toxicity: | 0.018 |
| Rat Oral Acute Toxicity: | 0.015 | Maximum Recommended Daily Dose: | 0.021 |
| Skin Sensitization: | 0.96 | Carcinogencity: | 0.042 |
| Eye Corrosion: | 0.034 | Eye Irritation: | 0.665 |
| Respiratory Toxicity: | 0.425 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC001553 |  |
0.865 | D0O1PH |  |
0.688 | ||
| ENC001678 |  |
0.831 | D07ILQ |  |
0.593 | ||
| ENC001646 |  |
0.826 | D00AOJ |  |
0.528 | ||
| ENC001602 |  |
0.826 | D00FGR |  |
0.469 | ||
| ENC001593 |  |
0.784 | D0O1TC | 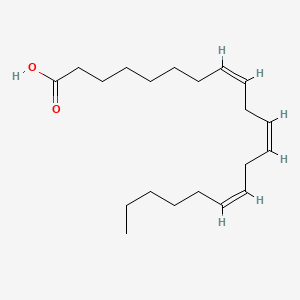 |
0.467 | ||
| ENC000608 | 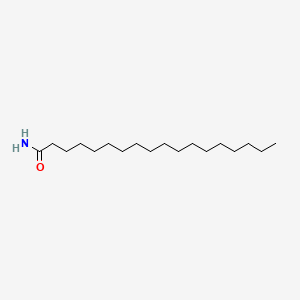 |
0.775 | D0Z5SM |  |
0.464 | ||
| ENC001708 | 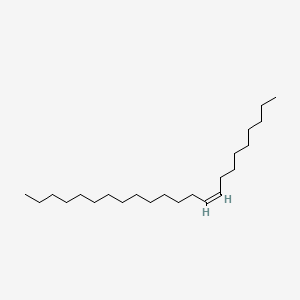 |
0.766 | D0OR6A | 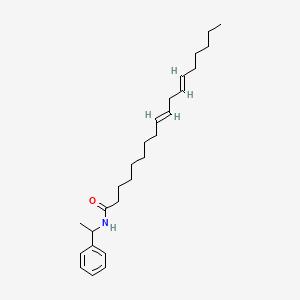 |
0.415 | ||
| ENC001689 |  |
0.757 | D0UE9X |  |
0.400 | ||
| ENC001627 |  |
0.753 | D05ATI |  |
0.398 | ||
| ENC002275 |  |
0.753 | D00STJ | 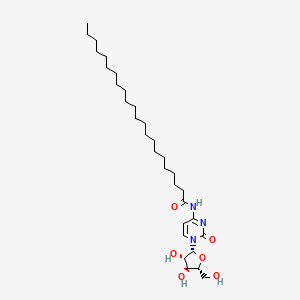 |
0.397 | ||