NPs Basic Information
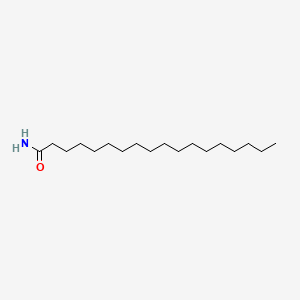
|
Name |
Octadecanamide
|
| Molecular Formula | C18H37NO | |
| IUPAC Name* |
octadecanamide
|
|
| SMILES |
CCCCCCCCCCCCCCCCCC(=O)N
|
|
| InChI |
InChI=1S/C18H37NO/c1-2-3-4-5-6-7-8-9-10-11-12-13-14-15-16-17-18(19)20/h2-17H2,1H3,(H2,19,20)
|
|
| InChIKey |
LYRFLYHAGKPMFH-UHFFFAOYSA-N
|
|
| Synonyms |
Octadecanamide; Stearamide; 124-26-5; STEARIC ACID AMIDE; Stearylamide; Octadecamide; Octadecylamide; Stearoylamide; Stearoylamine; Stearic amide; Kemamide S; Adogen 42; Crodamide S; Petrac vyn-eze; Amide T; NSC 66462; Amide C18; Armid 18; CHEBI:34900; Octadecanoic acid amide; YQX129FH1U; NSC-66462; CCRIS 6866; HSDB 723; EINECS 204-693-2; UNII-YQX129FH1U; BRN 0909006; AI3-10003; Crodamide SR; Crodamide S, SR; stearic acid, amide; ORISTAR STA; Amide C-18; STEARAMIDE [INCI]; EC 204-693-2; SCHEMBL27869; CHEMBL88311; DTXSID9027025; SCHEMBL20752935; STEARIC ACID AMIDE [HSDB]; NSC66462; ZINC8437471; BDBM50463974; LMFA08010003; MFCD00008038; STL453616; AKOS015843174; Octadecanamide, technical grade, 85%; HY-W130610; STEARAMIDE (STEARIC ACID AMIDE); NCGC00164016-01; BS-42312; CS-0196615; FT-0651996; EN300-83818; E82323; A805214; J-005061; J-523821; Q27116315; Z1245693822
|
|
| CAS | 124-26-5 | |
| PubChem CID | 31292 | |
| ChEMBL ID | CHEMBL88311 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 283.5 | ALogp: | 6.8 |
| HBD: | 1 | HBA: | 1 |
| Rotatable Bonds: | 16 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 43.1 | Aromatic Rings: | 0 |
| Heavy Atoms: | 20 | QED Weighted: | 0.354 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.872 | MDCK Permeability: | 0.00001750 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.002 | 20% Bioavailability (F20%): | 0.576 |
| 30% Bioavailability (F30%): | 0.997 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.234 | Plasma Protein Binding (PPB): | 98.06% |
| Volume Distribution (VD): | 1.409 | Fu: | 1.11% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.239 | CYP1A2-substrate: | 0.195 |
| CYP2C19-inhibitor: | 0.373 | CYP2C19-substrate: | 0.053 |
| CYP2C9-inhibitor: | 0.122 | CYP2C9-substrate: | 0.923 |
| CYP2D6-inhibitor: | 0.067 | CYP2D6-substrate: | 0.063 |
| CYP3A4-inhibitor: | 0.239 | CYP3A4-substrate: | 0.03 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 4.298 | Half-life (T1/2): | 0.084 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.428 | Human Hepatotoxicity (H-HT): | 0.019 |
| Drug-inuced Liver Injury (DILI): | 0.069 | AMES Toxicity: | 0.008 |
| Rat Oral Acute Toxicity: | 0.013 | Maximum Recommended Daily Dose: | 0.011 |
| Skin Sensitization: | 0.939 | Carcinogencity: | 0.043 |
| Eye Corrosion: | 0.213 | Eye Irritation: | 0.924 |
| Respiratory Toxicity: | 0.332 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000688 | 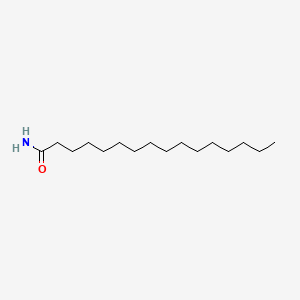 |
0.895 | D07ILQ |  |
0.696 | ||
| ENC000110 | 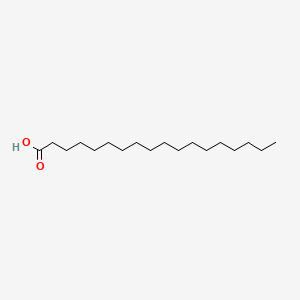 |
0.839 | D00AOJ | 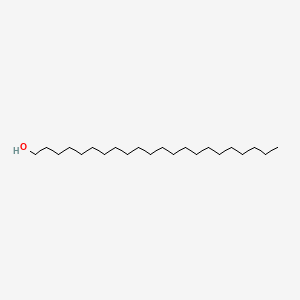 |
0.610 | ||
| ENC000280 | 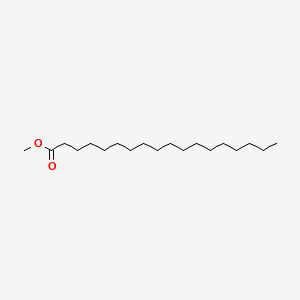 |
0.800 | D0Z5SM |  |
0.542 | ||
| ENC000356 | 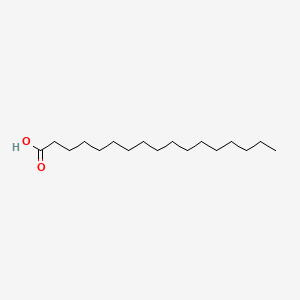 |
0.790 | D0O1PH |  |
0.538 | ||
| ENC001710 | 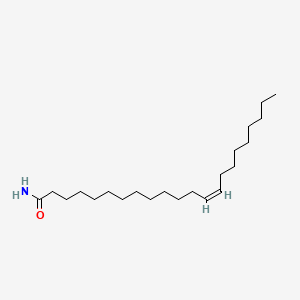 |
0.775 | D00FGR | 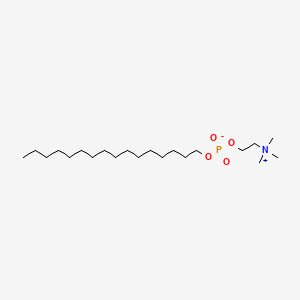 |
0.535 | ||
| ENC000497 |  |
0.765 | D05ATI |  |
0.465 | ||
| ENC000357 |  |
0.765 | D00STJ |  |
0.437 | ||
| ENC000258 |  |
0.765 | D0P1RL |  |
0.376 | ||
| ENC000400 |  |
0.758 | D0T9TJ |  |
0.372 | ||
| ENC000486 |  |
0.758 | D00MLW |  |
0.369 | ||