NPs Basic Information
|
Name |
Elaidamide
|
| Molecular Formula | C18H35NO | |
| IUPAC Name* |
(E)-octadec-9-enamide
|
|
| SMILES |
CCCCCCCC/C=C/CCCCCCCC(=O)N
|
|
| InChI |
InChI=1S/C18H35NO/c1-2-3-4-5-6-7-8-9-10-11-12-13-14-15-16-17-18(19)20/h9-10H,2-8,11-17H2,1H3,(H2,19,20)/b10-9+
|
|
| InChIKey |
FATBGEAMYMYZAF-MDZDMXLPSA-N
|
|
| Synonyms |
ELAIDAMIDE; 4303-70-2; 9-Octadecenamide; (E)-9-Octadecenamide; octadec-9-enamide; (E)-octadec-9-enamide; (9E)-9-Octadecenamide; 9E-hexadecenamide; elaidic acid amide; GOU8K597IT; (9E)-OCTADEC-9-ENAMIDE; UNII-GOU8K597IT; Armid ow; 9-Oleoamide; EINECS 224-316-5; trans-9-octadecenamide; 9-Octadecenamide, (9E)-; SCHEMBL19788; CHEMBL86554; SCHEMBL195221; CHEBI:165592; DTXSID901017170; (E)-9,10-OCTADECENAMIDE; LMFA08010011; ZINC14880924; AKOS000277608; DB03784; BS-16886; CS-0166850; D83824; Q27094678
|
|
| CAS | 4303-70-2 | |
| PubChem CID | 5353370 | |
| ChEMBL ID | CHEMBL86554 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 281.5 | ALogp: | 6.6 |
| HBD: | 1 | HBA: | 1 |
| Rotatable Bonds: | 15 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 43.1 | Aromatic Rings: | 0 |
| Heavy Atoms: | 20 | QED Weighted: | 0.303 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.804 | MDCK Permeability: | 0.00002710 |
| Pgp-inhibitor: | 0.002 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.006 | 20% Bioavailability (F20%): | 0.838 |
| 30% Bioavailability (F30%): | 0.993 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.447 | Plasma Protein Binding (PPB): | 99.66% |
| Volume Distribution (VD): | 3.035 | Fu: | 0.75% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.367 | CYP1A2-substrate: | 0.202 |
| CYP2C19-inhibitor: | 0.412 | CYP2C19-substrate: | 0.054 |
| CYP2C9-inhibitor: | 0.227 | CYP2C9-substrate: | 0.941 |
| CYP2D6-inhibitor: | 0.222 | CYP2D6-substrate: | 0.199 |
| CYP3A4-inhibitor: | 0.375 | CYP3A4-substrate: | 0.034 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 3.191 | Half-life (T1/2): | 0.28 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.23 | Human Hepatotoxicity (H-HT): | 0.032 |
| Drug-inuced Liver Injury (DILI): | 0.025 | AMES Toxicity: | 0.003 |
| Rat Oral Acute Toxicity: | 0.01 | Maximum Recommended Daily Dose: | 0.029 |
| Skin Sensitization: | 0.942 | Carcinogencity: | 0.035 |
| Eye Corrosion: | 0.094 | Eye Irritation: | 0.877 |
| Respiratory Toxicity: | 0.099 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
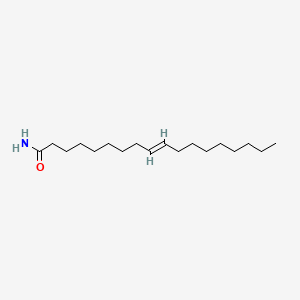
| ENC001602 | 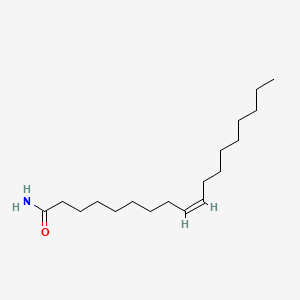 |
1.000 | D0O1PH |  |
0.757 | ||
| ENC002845 | 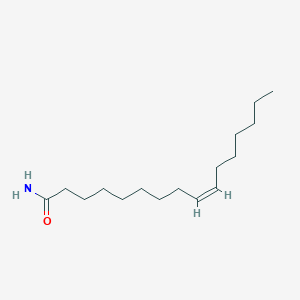 |
0.895 | D0O1TC | 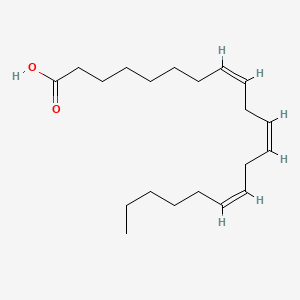 |
0.538 | ||
| ENC001555 | 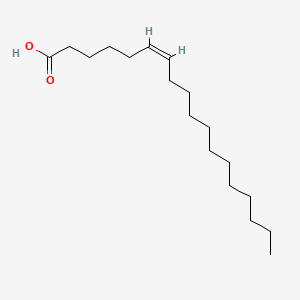 |
0.839 | D07ILQ |  |
0.519 | ||
| ENC001699 | 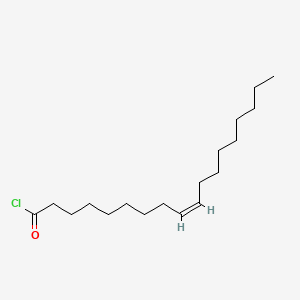 |
0.839 | D0OR6A | 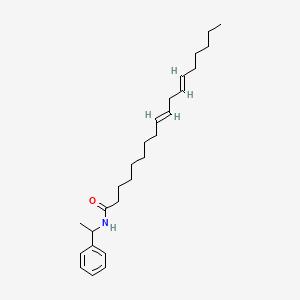 |
0.468 | ||
| ENC001710 | 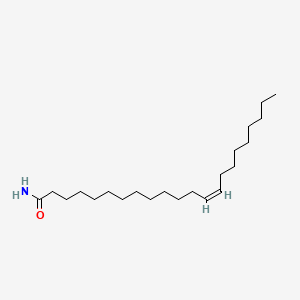 |
0.826 | D0UE9X | 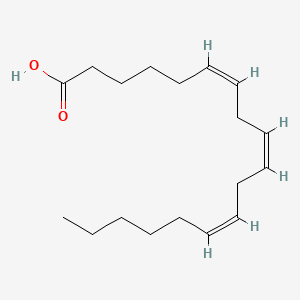 |
0.462 | ||
| ENC001688 | 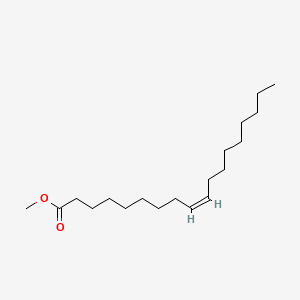 |
0.800 | D0Z5SM |  |
0.461 | ||
| ENC001775 | 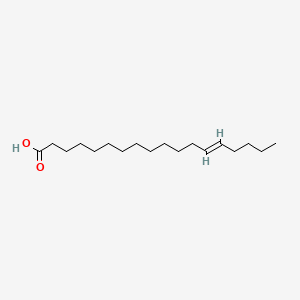 |
0.781 | D05ATI |  |
0.444 | ||
| ENC001593 | 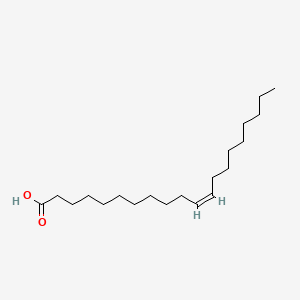 |
0.765 | D00AOJ |  |
0.393 | ||
| ENC001670 |  |
0.765 | D0Z5BC |  |
0.388 | ||
| ENC001687 |  |
0.746 | D09SRR | 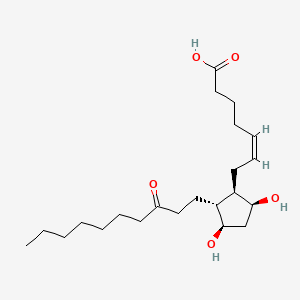 |
0.375 | ||