NPs Basic Information
|
Name |
Monoelaidin
|
| Molecular Formula | C21H40O4 | |
| IUPAC Name* |
2,3-dihydroxypropyl (E)-octadec-9-enoate
|
|
| SMILES |
CCCCCCCC/C=C/CCCCCCCC(=O)OCC(CO)O
|
|
| InChI |
InChI=1S/C21H40O4/c1-2-3-4-5-6-7-8-9-10-11-12-13-14-15-16-17-21(24)25-19-20(23)18-22/h9-10,20,22-23H,2-8,11-19H2,1H3/b10-9+
|
|
| InChIKey |
RZRNAYUHWVFMIP-MDZDMXLPSA-N
|
|
| Synonyms |
Monoelaidin; 2716-53-2; 1-monoelaidin; 2,3-dihydroxypropyl (E)-octadec-9-enoate; 2,3-Dihydroxypropyl elaidate; 9-Octadecenoic acid, 2,3-dihydroxypropyl ester, (E)-; 29798-65-0; 1-[(9E)-octadecenoyl]glycerol; 9-Octadecenoicacid(E)-,2,3-dihydroxypropylester; oleoyl-glycerol; 25496-72-4; trans-Monoelaidin; 1-elaidoylglycerol; glycerol mono-oleate; Glycerol alpha-Monoelaidate; SCHEMBL15604; Glycerol 1-(9-octadecenoate); SCHEMBL9754698; DTXSID0052831; CHEBI:131342; 1-[(E)-octadec-9-enoyl]glycerol; 251983-54-7; CAA71653; 1-[(9E)-octadec-9-enoyl]glycerol; MFCD00069635; 1-Mono-trans-9-octadecenoyl Glycerol; AKOS015894308; 2,3-dihydroxypropyl (9E)-octadec-9-enoate; 9-Octadecenoicacid-,2,3-dihydroxypropylester; M1075; 2,3-Dihydroxypropyl (9E)-9-octadecenoate #; E73975; J-016682; Q27225105
|
|
| CAS | 25496-72-4 | |
| PubChem CID | 5364833 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 356.5 | ALogp: | 6.5 |
| HBD: | 2 | HBA: | 4 |
| Rotatable Bonds: | 19 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 66.8 | Aromatic Rings: | 0 |
| Heavy Atoms: | 25 | QED Weighted: | 0.199 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.862 | MDCK Permeability: | 0.00004020 |
| Pgp-inhibitor: | 0.046 | Pgp-substrate: | 0.688 |
| Human Intestinal Absorption (HIA): | 0.035 | 20% Bioavailability (F20%): | 0.894 |
| 30% Bioavailability (F30%): | 0.979 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.344 | Plasma Protein Binding (PPB): | 98.96% |
| Volume Distribution (VD): | 0.76 | Fu: | 0.91% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.42 | CYP1A2-substrate: | 0.159 |
| CYP2C19-inhibitor: | 0.288 | CYP2C19-substrate: | 0.053 |
| CYP2C9-inhibitor: | 0.335 | CYP2C9-substrate: | 0.945 |
| CYP2D6-inhibitor: | 0.056 | CYP2D6-substrate: | 0.081 |
| CYP3A4-inhibitor: | 0.579 | CYP3A4-substrate: | 0.063 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 4.552 | Half-life (T1/2): | 0.791 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.172 | Human Hepatotoxicity (H-HT): | 0.034 |
| Drug-inuced Liver Injury (DILI): | 0.017 | AMES Toxicity: | 0.005 |
| Rat Oral Acute Toxicity: | 0.016 | Maximum Recommended Daily Dose: | 0.017 |
| Skin Sensitization: | 0.94 | Carcinogencity: | 0.067 |
| Eye Corrosion: | 0.005 | Eye Irritation: | 0.091 |
| Respiratory Toxicity: | 0.249 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
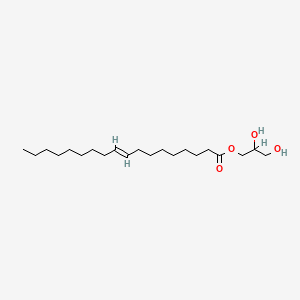
| ENC001930 | 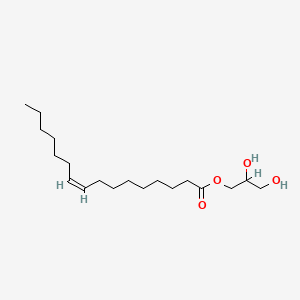 |
0.915 | D0O1PH |  |
0.713 | ||
| ENC001628 | 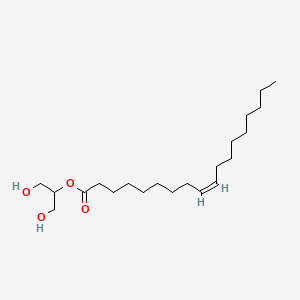 |
0.775 | D07ILQ |  |
0.506 | ||
| ENC001643 |  |
0.772 | D0O1TC |  |
0.473 | ||
| ENC000484 |  |
0.766 | D00MLW |  |
0.422 | ||
| ENC001670 |  |
0.763 | D0OR6A | 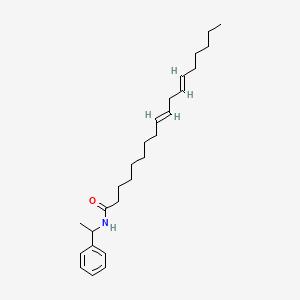 |
0.421 | ||
| ENC001679 | 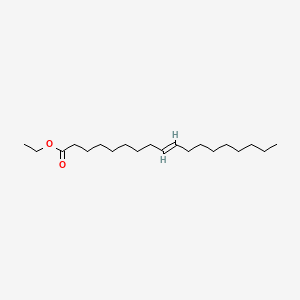 |
0.763 | D0UE9X | 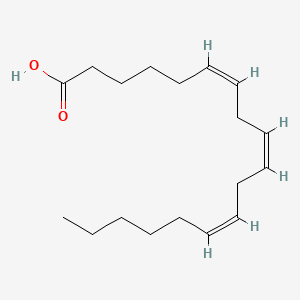 |
0.407 | ||
| ENC005019 | 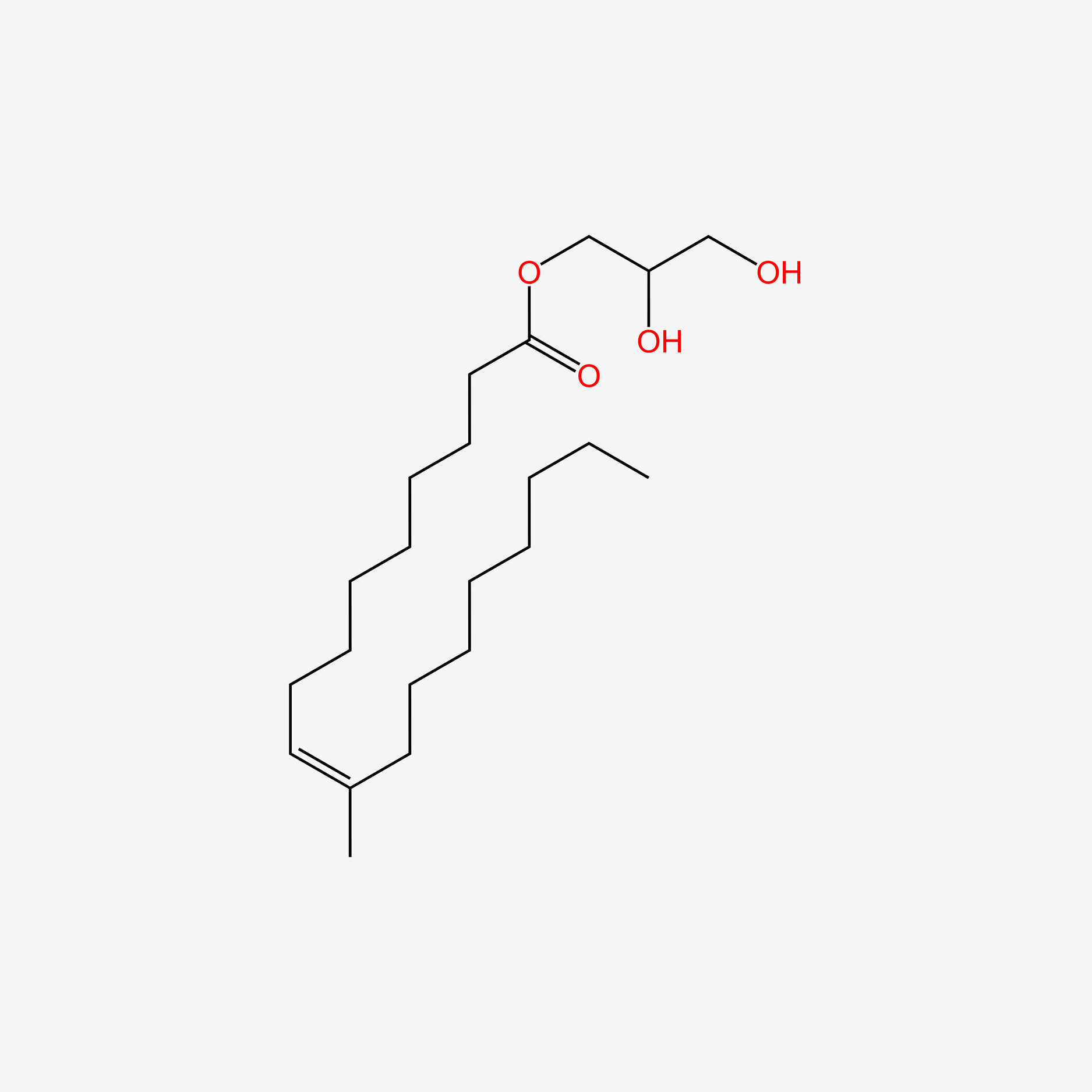 |
0.756 | D0Z5SM |  |
0.404 | ||
| ENC000572 |  |
0.724 | D00AOJ |  |
0.394 | ||
| ENC001657 |  |
0.724 | D05ATI |  |
0.388 | ||
| ENC001680 |  |
0.724 | D0H2YX | 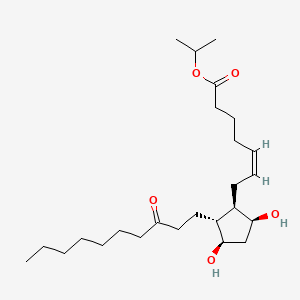 |
0.375 | ||