NPs Basic Information
|
Name |
gamma-Oryzanol
|
| Molecular Formula | C40H58O4 | |
| IUPAC Name* |
[(1S,3R,6S,8R,11S,12S,15R,16R)-7,7,12,16-tetramethyl-15-[(2R)-6-methylhept-5-en-2-yl]-6-pentacyclo[9.7.0.01,3.03,8.012,16]octadecanyl] (E)-3-(4-hydroxy-3-methoxyphenyl)prop-2-enoate
|
|
| SMILES |
C[C@H](CCC=C(C)C)[C@H]1CC[C@@]2([C@@]1(CC[C@]34[C@H]2CC[C@@H]5[C@]3(C4)CC[C@@H](C5(C)C)OC(=O)/C=C/C6=CC(=C(C=C6)O)OC)C)C
|
|
| InChI |
InChI=1S/C40H58O4/c1-26(2)10-9-11-27(3)29-18-20-38(7)33-16-15-32-36(4,5)34(19-21-39(32)25-40(33,39)23-22-37(29,38)6)44-35(42)17-13-28-12-14-30(41)31(24-28)43-8/h10,12-14,17,24,27,29,32-34,41H,9,11,15-16,18-23,25H2,1-8H3/b17-13+/t27-,29-,32+,33+,34+,37-,38+,39-,40+/m1/s1
|
|
| InChIKey |
FODTZLFLDFKIQH-FSVGXZBPSA-N
|
|
| Synonyms |
gamma-Oryzanol; Cycloartenyl ferulate; gamma Oryzanol; Oliver; 11042-64-1; Gammariza; gamma-Orizanol; HI-Z; Cycloartenol Trans-Ferulate; Cycloartenol ferulate; Gamma oryzanol [JAN]; 3-O-Ferulylcycloartenol; 3PQR2YON9T; Ferulic acid cycloartenol ester; Oryzanol A; gamma Oryzanol (JAN); DSSTox_CID_1084; 21238-33-5; DSSTox_RID_75932; DSSTox_GSID_21084; CAS-11042-64-1; UNII-SST9XCL51M; Cycloartenol ferulic acid ester; CCRIS 4251; y-Oryzanol; gamma -Oryzanol; NCGC00181756-01; 9,19-CYCLOLANOST-24-EN-3-OL, 3-((2E)-3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOATE), (3.BETA.)-; 9,19-Cyclolanost-24-en-3-ol, 3-[(2E)-3-(4-hydroxy-3-methoxyphenyl)-2-propenoate], (3.beta.)-; gamma-Oryzanol (TN); ORYZANOL A [MI]; UNII-3PQR2YON9T; SST9XCL51M; Hi-Z (TN); Cycloartenol, trans-ferulate; SCHEMBL718726; (3beta)-9,19-Cyclolanost-24-en-3-yl 4-hydroxy-3-methoxycinnamate; CHEMBL388595; CHEBI:32345; CYCLOARTENYL FERULATE [INCI]; EINECS 244-285-1; Tox21_112482; BDBM50483936; MFCD00867548; s3957; ZINC28646872; AKOS016339566; Tox21_112482_1; CCG-270201; OZ; HY-125938; CS-0103198; D01221; E88692; J-002421; Q27257875; 9,19-Cyclo-9beta-lanost-24-en-3beta-ol 4-hydroxy-3-methoxycinamate; 9,19-CYCLO-9.BETA.-LANOST-24-EN-3.BETA.-OL 4-HYDROXY-3-METHOXYCINAMATE; 9,19-CYCLO-9.BETA.-LANOST-24-EN-3.BETA.-OL, 4-HYDROXY-3-METHOXYCINNAMATE; [(1S,3R,6S,8R,11S,12S,15R,16R)-7,7,12,16-tetramethyl-15-[(2R)-6-methylhept-5-en-2-yl]-6-pentacyclo[9.7.0.01,3.03,8.012,16]octadecanyl] (E)-3-(4-hydroxy-3-methoxyphenyl)prop-2-enoate; 286011-27-6; 9,19-CYCLOLANOST-24-EN-3-OL, (2E)-3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOATE, (3.BETA.)-; 9,19-CYCLOLANOST-24-EN-3-OL, 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOATE, (3.BETA.)-; CINNAMIC ACID, 4-HYDROXY-3-METHOXY-, 9,19-CYCLO-9.BETA.-LANOST-24-EN-3.BETA.-YL ESTER
|
|
| CAS | 11042-64-1 | |
| PubChem CID | 5282164 | |
| ChEMBL ID | CHEMBL388595 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 602.9 | ALogp: | 12.1 |
| HBD: | 1 | HBA: | 4 |
| Rotatable Bonds: | 9 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 55.8 | Aromatic Rings: | 6 |
| Heavy Atoms: | 44 | QED Weighted: | 0.151 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.044 | MDCK Permeability: | 0.00001930 |
| Pgp-inhibitor: | 0.978 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.003 | 20% Bioavailability (F20%): | 0.945 |
| 30% Bioavailability (F30%): | 0.985 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.013 | Plasma Protein Binding (PPB): | 97.37% |
| Volume Distribution (VD): | 2.409 | Fu: | 2.72% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.045 | CYP1A2-substrate: | 0.26 |
| CYP2C19-inhibitor: | 0.261 | CYP2C19-substrate: | 0.906 |
| CYP2C9-inhibitor: | 0.169 | CYP2C9-substrate: | 0.907 |
| CYP2D6-inhibitor: | 0.568 | CYP2D6-substrate: | 0.825 |
| CYP3A4-inhibitor: | 0.757 | CYP3A4-substrate: | 0.747 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 8.742 | Half-life (T1/2): | 0.04 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.949 | Human Hepatotoxicity (H-HT): | 0.482 |
| Drug-inuced Liver Injury (DILI): | 0.198 | AMES Toxicity: | 0.002 |
| Rat Oral Acute Toxicity: | 0.101 | Maximum Recommended Daily Dose: | 0.872 |
| Skin Sensitization: | 0.969 | Carcinogencity: | 0.04 |
| Eye Corrosion: | 0.014 | Eye Irritation: | 0.084 |
| Respiratory Toxicity: | 0.868 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
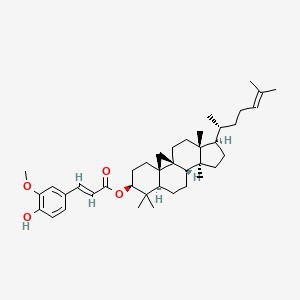
| ENC001745 | 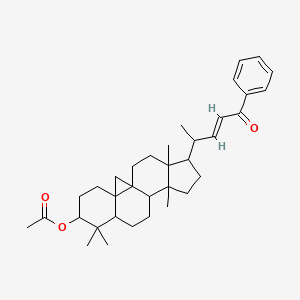 |
0.551 | D0X7XG | 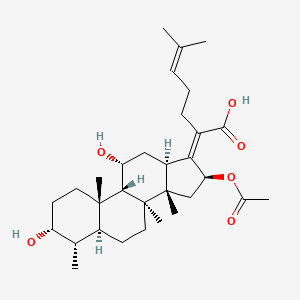 |
0.277 | ||
| ENC002119 | 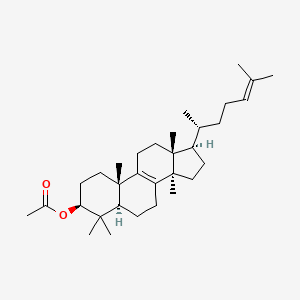 |
0.466 | D03MTN |  |
0.252 | ||
| ENC001942 |  |
0.440 | D0U0XD |  |
0.241 | ||
| ENC002686 |  |
0.399 | D07VBA | 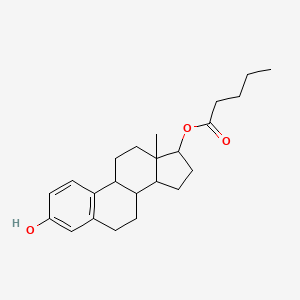 |
0.241 | ||
| ENC001833 | 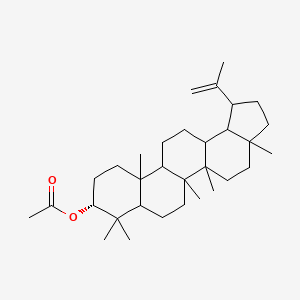 |
0.381 | D0Y7LD | 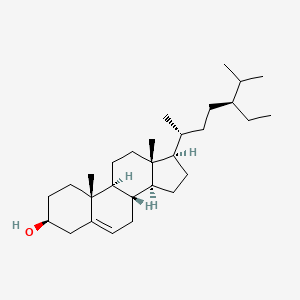 |
0.235 | ||
| ENC005285 | 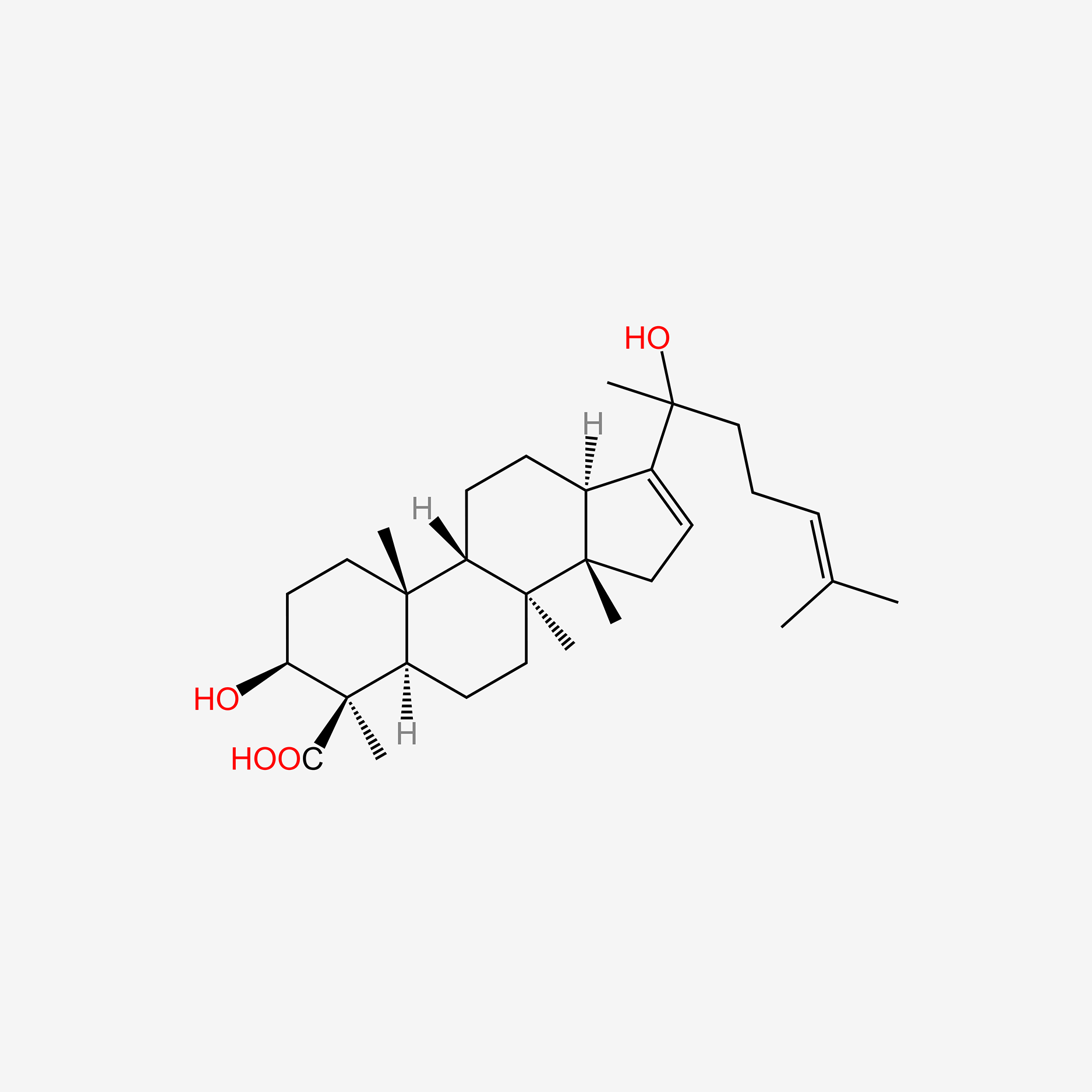 |
0.357 | D06AWE | 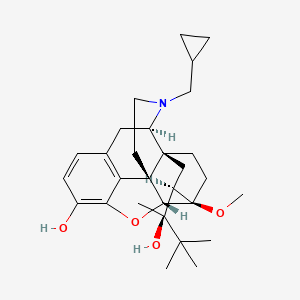 |
0.234 | ||
| ENC006069 | 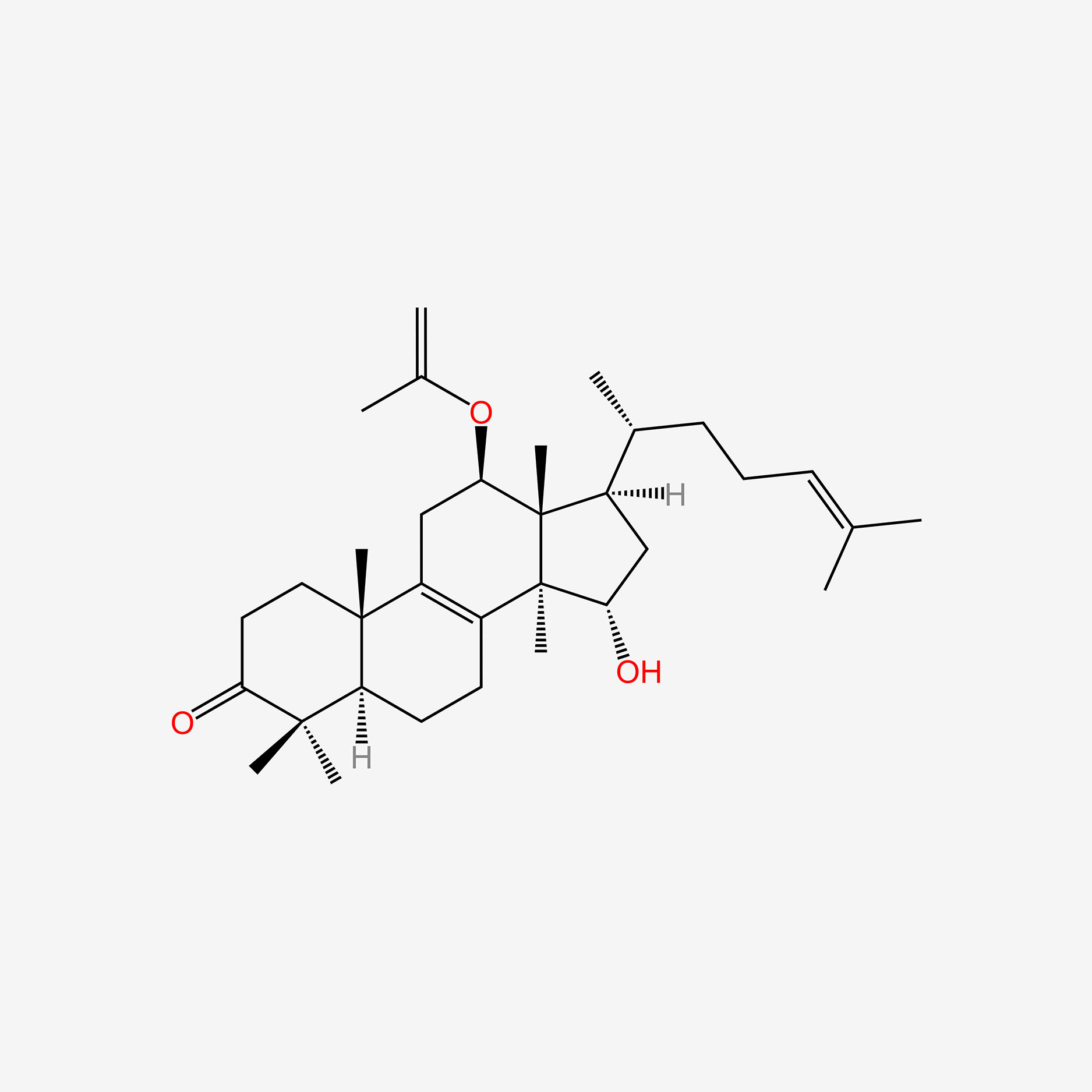 |
0.354 | D0H2JP | 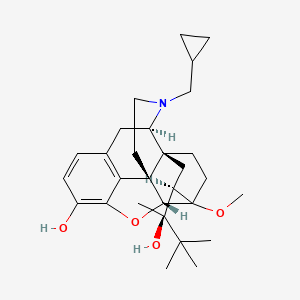 |
0.234 | ||
| ENC006068 | 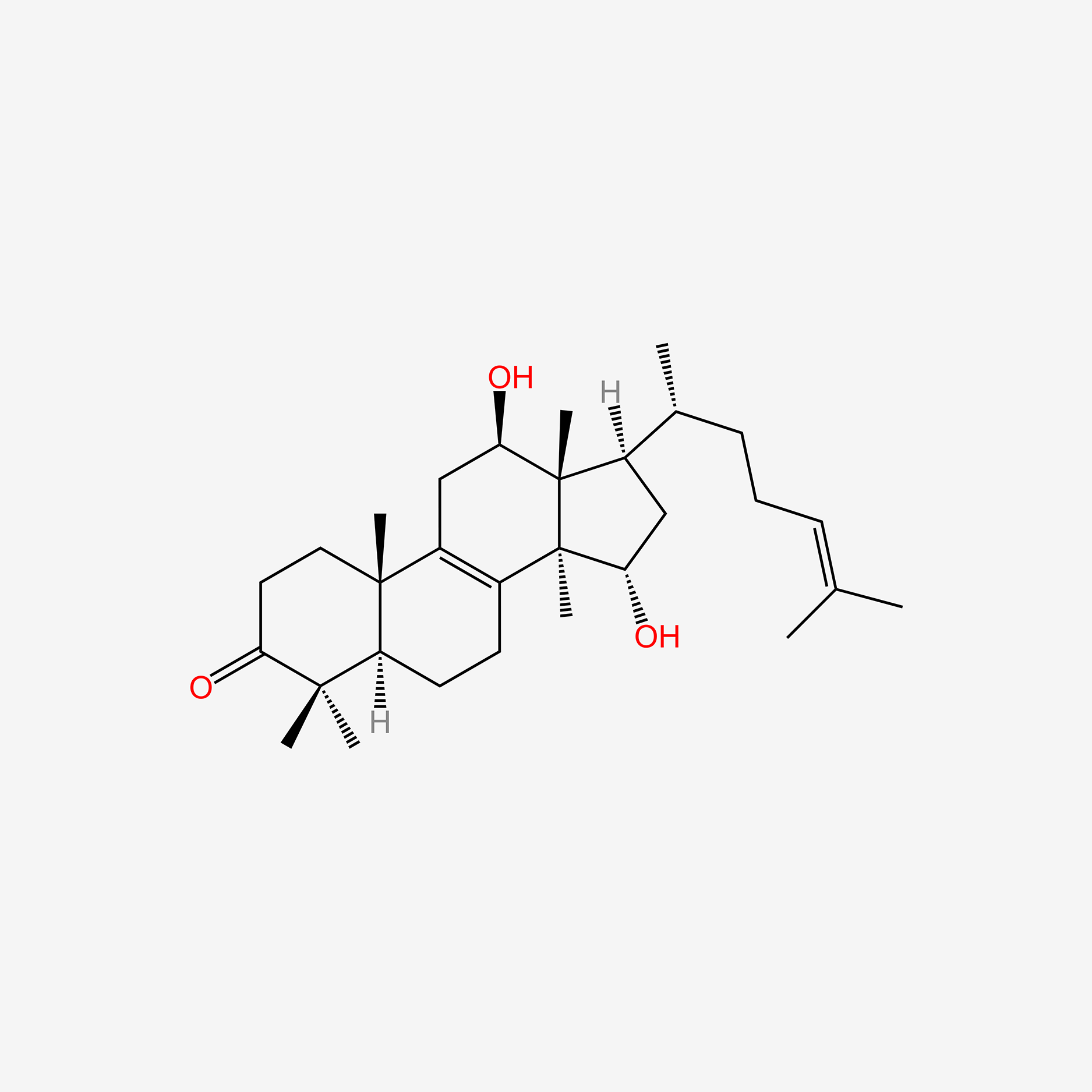 |
0.338 | D08TEJ | 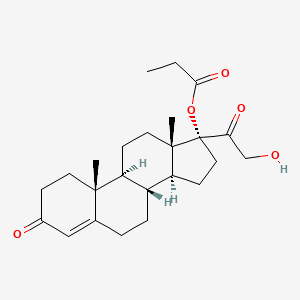 |
0.232 | ||
| ENC003874 | 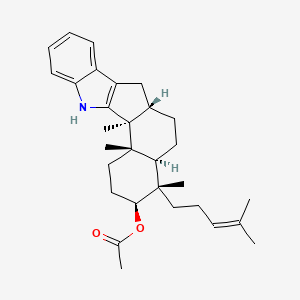 |
0.338 | D00AEQ | 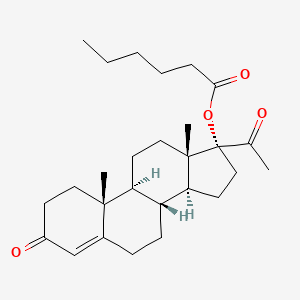 |
0.231 | ||
| ENC002012 | 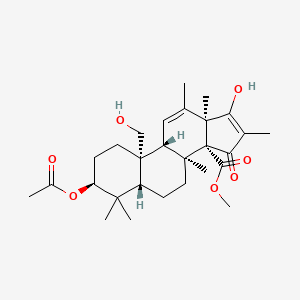 |
0.321 | D0G3SH | 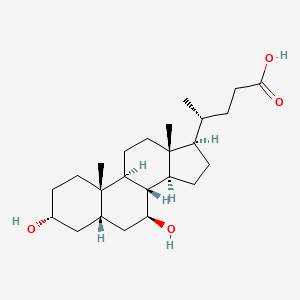 |
0.228 | ||