NPs Basic Information
|
Name |
Esculetin
|
| Molecular Formula | C9H6O4 | |
| IUPAC Name* |
6,7-dihydroxychromen-2-one
|
|
| SMILES |
C1=CC(=O)OC2=CC(=C(C=C21)O)O
|
|
| InChI |
InChI=1S/C9H6O4/c10-6-3-5-1-2-9(12)13-8(5)4-7(6)11/h1-4,10-11H
|
|
| InChIKey |
ILEDWLMCKZNDJK-UHFFFAOYSA-N
|
|
| Synonyms |
Esculetin; 305-01-1; 6,7-DIHYDROXYCOUMARIN; Aesculetin; 6,7-Dihydroxy-2H-chromen-2-one; Cichorigenin; Esculetol; Cichoriin aglucon; Esculatin; Esculin aglucon; Esculin aglycon; Cichoriin aglycon; Asculetine; 6,7-dihydroxychromen-2-one; 2H-1-Benzopyran-2-one, 6,7-dihydroxy-; 6,7-Dihydroxy-2-benzopyrone; 6,7-Dihydroxy-2H-1-benzopyran-2-one; Coumarin, 6,7-dihydroxy-; NSC 26428; 91753-33-2; 6,7-bis(oxidanyl)chromen-2-one; 6,7-Dihydroxycounmarin; MFCD00006874; MLS000069479; CHEMBL244743; SM2XD6V944; CHEBI:490095; 6,7-dihydroxy-1-benzopyran-2-one; NSC26428; NSC-26428; SMR000059055; esculetine; CCRIS 7065; EINECS 206-161-5; BRN 0152788; UNII-SM2XD6V944; Esculetol); Aesculetin ,(S); Cichorigenin|Aesculetin; Spectrum_001166; Aesculetin (cichorigenin; 6,7-dihydroxy-coumarin; SpecPlus_000334; ESCULETIN [MI]; ESCULETIN [INCI]; Prestwick0_000940; Prestwick1_000940; Prestwick2_000940; Prestwick3_000940; Spectrum2_000586; Spectrum3_000752; Spectrum4_001886; Spectrum5_000512; bmse000986; Oprea1_719746; SCHEMBL24641; 6,7-Dihydroxy-2-chromenone; BSPBio_000880; BSPBio_002364; KBioGR_002416; KBioSS_001646; 5-18-03-00202 (Beilstein Handbook Reference); 6,7-Dihydroxycoumarin, 8CI; DivK1c_006430; Esculetin, analytical standard; SPECTRUM1500899; SPBio_000432; SPBio_003049; 6,7-Dihydroxycoumarin, 98%; BPBio1_000968; GTPL5180; DTXSID3075383; BDBM34571; cid_5281416; KBio1_001374; KBio2_001646; KBio2_004214; KBio2_006782; KBio3_001584; ZINC57908; DTXSID801293090; HMS1570L22; HMS1921M14; HMS2097L22; HMS2233G24; HMS3373K09; KUC108669N; 2,6-Dihydroxy-7H-chromen-7-one; ALBB-023362; AMY25634; EX-A6796; HY-N0284; Coumarin, 6,7-dihydroxy- Esculetin; BBL037229; CCG-38509; s4711; STL560155; 6,7-Dihydroxy-2H-chromen-2-one #; AKOS000276955; Esculetin [Matrix for MALDI-TOF/MS]; SDCCGMLS-0066669.P001; 2,6-Dihydroxy-7H-1-benzopyran-7-one; SMP2_000093; NCGC00016425-01; NCGC00016425-02; NCGC00016425-03; NCGC00016425-04; NCGC00016425-05; NCGC00016425-06; NCGC00094873-01; NCGC00094873-02; AC-18400; AC-34573; AS-11863; CAS-305-01-1; KSC-11-243-1; NCI60_002119; SY036794; DB-014626; AB00489955; CS-0008780; E0386; E1287; FT-0620854; A14553; C09263; E-3400; E-3401; O11524; 2H-1-Benzopyran-2-one, 6,7-dihydroxy- (9CI); 305E011; A820407; SR-01000721907; Q3058018; SR-01000721907-2; BRD-K58149231-001-06-9; BRD-K58149231-001-10-1; B0005-464319; Esculetin, European Pharmacopoeia (EP) Reference Standard; HFC
|
|
| CAS | 305-01-1 | |
| PubChem CID | 5281416 | |
| ChEMBL ID | CHEMBL244743 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 178.14 | ALogp: | 1.2 |
| HBD: | 2 | HBA: | 4 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 66.8 | Aromatic Rings: | 2 |
| Heavy Atoms: | 13 | QED Weighted: | 0.475 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.765 | MDCK Permeability: | 0.00001240 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.937 |
| Human Intestinal Absorption (HIA): | 0.012 | 20% Bioavailability (F20%): | 0.997 |
| 30% Bioavailability (F30%): | 1 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.031 | Plasma Protein Binding (PPB): | 85.21% |
| Volume Distribution (VD): | 0.487 | Fu: | 19.83% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.91 | CYP1A2-substrate: | 0.74 |
| CYP2C19-inhibitor: | 0.053 | CYP2C19-substrate: | 0.056 |
| CYP2C9-inhibitor: | 0.061 | CYP2C9-substrate: | 0.757 |
| CYP2D6-inhibitor: | 0.472 | CYP2D6-substrate: | 0.609 |
| CYP3A4-inhibitor: | 0.434 | CYP3A4-substrate: | 0.151 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 16.579 | Half-life (T1/2): | 0.882 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.049 | Human Hepatotoxicity (H-HT): | 0.116 |
| Drug-inuced Liver Injury (DILI): | 0.882 | AMES Toxicity: | 0.12 |
| Rat Oral Acute Toxicity: | 0.099 | Maximum Recommended Daily Dose: | 0.561 |
| Skin Sensitization: | 0.921 | Carcinogencity: | 0.721 |
| Eye Corrosion: | 0.395 | Eye Irritation: | 0.979 |
| Respiratory Toxicity: | 0.301 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
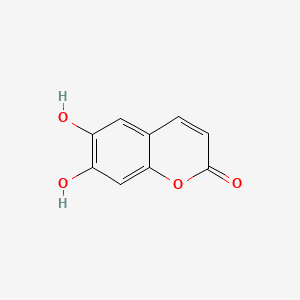
| ENC001537 | 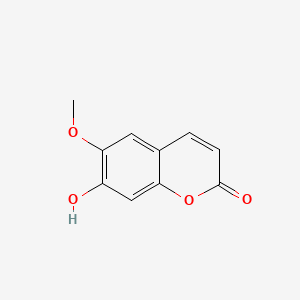 |
0.667 | D04AIT |  |
0.362 | ||
| ENC001562 | 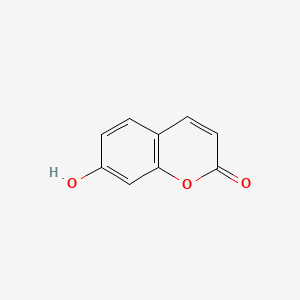 |
0.556 | D0K8KX | 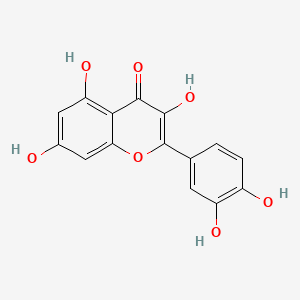 |
0.352 | ||
| ENC001524 | 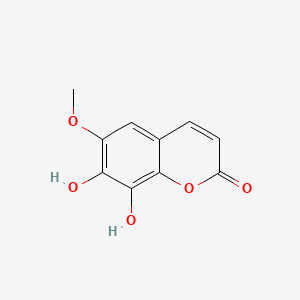 |
0.453 | D08SKH | 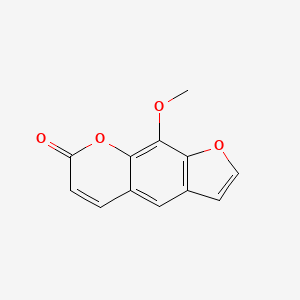 |
0.344 | ||
| ENC001539 | 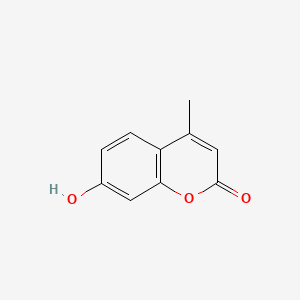 |
0.440 | D0T7OW | 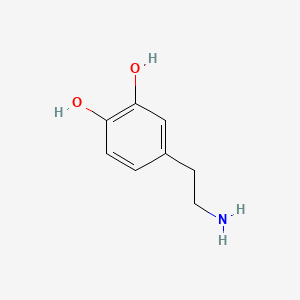 |
0.320 | ||
| ENC002806 | 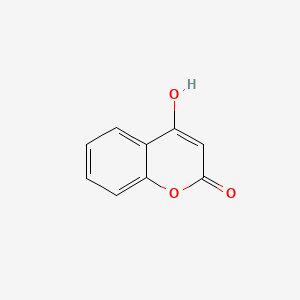 |
0.429 | D0V9EN | 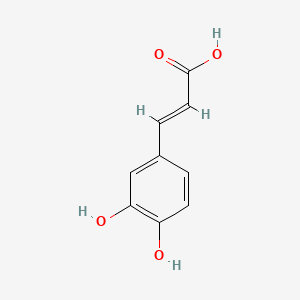 |
0.315 | ||
| ENC000025 | 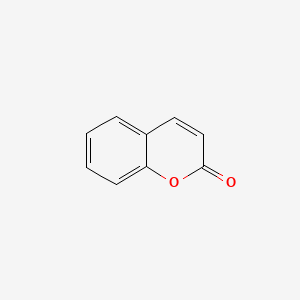 |
0.417 | D07MOX | 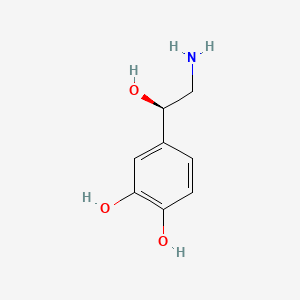 |
0.308 | ||
| ENC004389 | 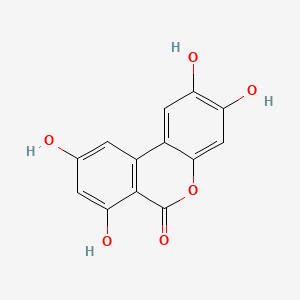 |
0.397 | D08HVR | 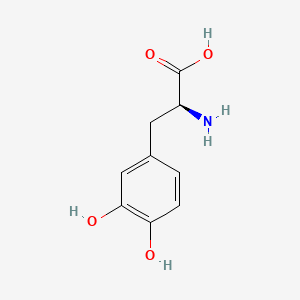 |
0.304 | ||
| ENC001747 | 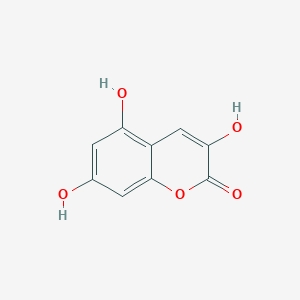 |
0.396 | D06GCK |  |
0.296 | ||
| ENC003772 | 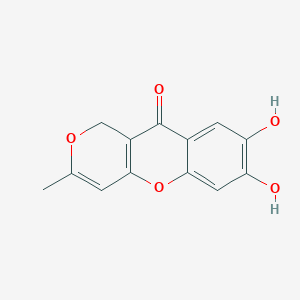 |
0.387 | D07MGA |  |
0.293 | ||
| ENC001447 | 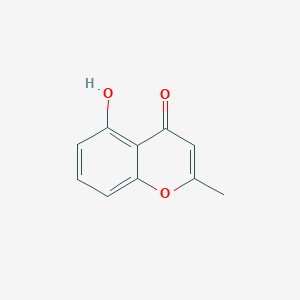 |
0.385 | D0BA6T | 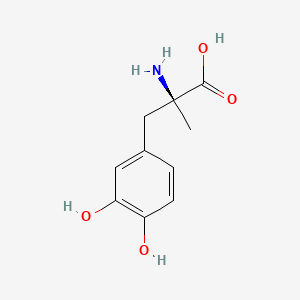 |
0.293 | ||