NPs Basic Information
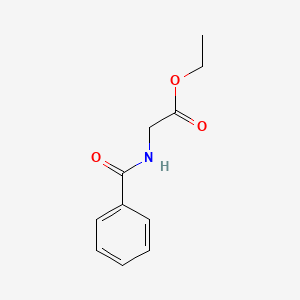
|
Name |
Ethyl Hippurate
|
| Molecular Formula | C11H13NO3 | |
| IUPAC Name* |
ethyl 2-benzamidoacetate
|
|
| SMILES |
CCOC(=O)CNC(=O)C1=CC=CC=C1
|
|
| InChI |
InChI=1S/C11H13NO3/c1-2-15-10(13)8-12-11(14)9-6-4-3-5-7-9/h3-7H,2,8H2,1H3,(H,12,14)
|
|
| InChIKey |
PTXRQIPIELXJFH-UHFFFAOYSA-N
|
|
| Synonyms |
Ethyl Hippurate; 1499-53-2; ethyl 2-benzamidoacetate; N-Benzoylglycine ethyl ester; ethyl 2-(benzoylamino)acetate; ethyl 2-(phenylformamido)acetate; ethyl N-benzoylglycinate; Benzoic amide, N-(ethoxycarbonyl)methyl-; Ethylhippurate; Glycine, N-benzoyl-, ethyl ester; ethyl benzoylaminoacetate; Maybridge1_006087; Ethyl (benzoylamino)acetate; CBDivE_004878; MLS000532642; Ethyl (benzoylamino)acetate #; CCG-10; SCHEMBL1221369; CHEMBL1558767; BENZOYLGLYCINE ETHYL ESTER; HMS558M15; DTXSID50280491; ethyl N-(phenylcarbonyl)glycinate; benzoylaminoacetic acid ethyl ester; HMS2168B06; HMS3311J13; BAA49953; NSC17140; NSC17495; ZINC1748026; Benzoylamino-acetic acid ethyl ester; MFCD00026890; NSC-17140; NSC-17495; STK123002; AKOS000496044; SMR000137581; DB-043029; CS-0206786; FT-0633614; 1M-011
|
|
| CAS | 1499-53-2 | |
| PubChem CID | 226558 | |
| ChEMBL ID | CHEMBL1558767 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 207.23 | ALogp: | 1.2 |
| HBD: | 1 | HBA: | 3 |
| Rotatable Bonds: | 5 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 55.4 | Aromatic Rings: | 1 |
| Heavy Atoms: | 15 | QED Weighted: | 0.759 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.422 | MDCK Permeability: | 0.00005220 |
| Pgp-inhibitor: | 0.002 | Pgp-substrate: | 0.002 |
| Human Intestinal Absorption (HIA): | 0.006 | 20% Bioavailability (F20%): | 0.002 |
| 30% Bioavailability (F30%): | 0.346 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.847 | Plasma Protein Binding (PPB): | 49.45% |
| Volume Distribution (VD): | 0.635 | Fu: | 37.36% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.895 | CYP1A2-substrate: | 0.081 |
| CYP2C19-inhibitor: | 0.394 | CYP2C19-substrate: | 0.263 |
| CYP2C9-inhibitor: | 0.144 | CYP2C9-substrate: | 0.464 |
| CYP2D6-inhibitor: | 0.016 | CYP2D6-substrate: | 0.235 |
| CYP3A4-inhibitor: | 0.028 | CYP3A4-substrate: | 0.223 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 7.285 | Half-life (T1/2): | 0.858 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.199 | Human Hepatotoxicity (H-HT): | 0.034 |
| Drug-inuced Liver Injury (DILI): | 0.509 | AMES Toxicity: | 0.008 |
| Rat Oral Acute Toxicity: | 0.013 | Maximum Recommended Daily Dose: | 0.008 |
| Skin Sensitization: | 0.238 | Carcinogencity: | 0.033 |
| Eye Corrosion: | 0.006 | Eye Irritation: | 0.155 |
| Respiratory Toxicity: | 0.043 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000175 | 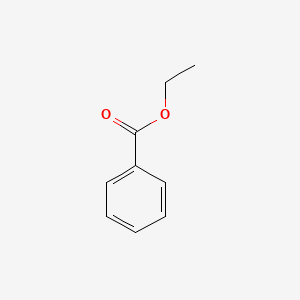 |
0.587 | D0X9RY | 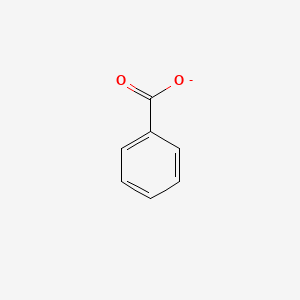 |
0.426 | ||
| ENC001397 | 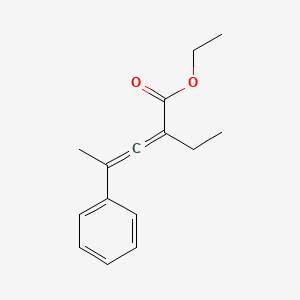 |
0.483 | D0B7OD | 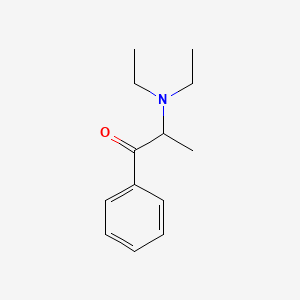 |
0.407 | ||
| ENC000596 | 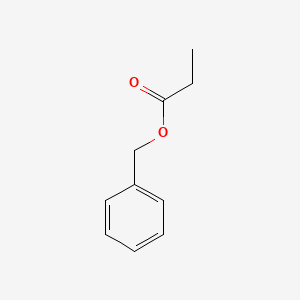 |
0.462 | D0G2MH | 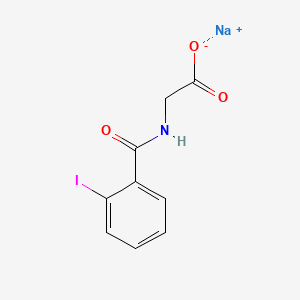 |
0.397 | ||
| ENC000174 | 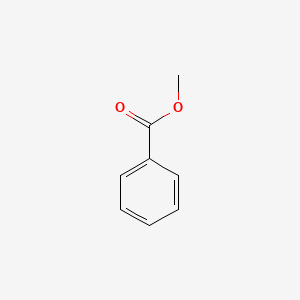 |
0.458 | D05OFX | 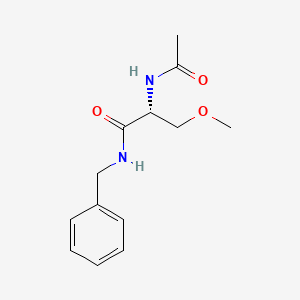 |
0.394 | ||
| ENC000192 | 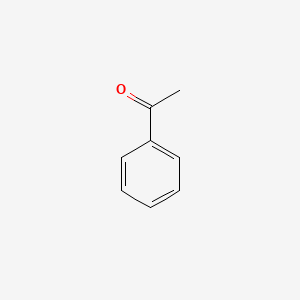 |
0.457 | D07ONP | 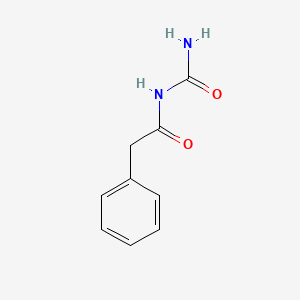 |
0.393 | ||
| ENC000176 | 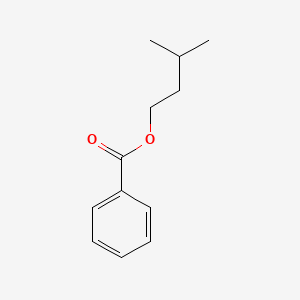 |
0.446 | D06OAV |  |
0.373 | ||
| ENC000637 |  |
0.444 | D05KON |  |
0.369 | ||
| ENC001726 |  |
0.441 | D00DZN |  |
0.362 | ||
| ENC001151 | 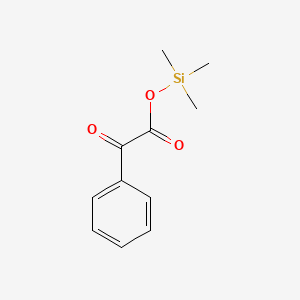 |
0.439 | D0S7VO | 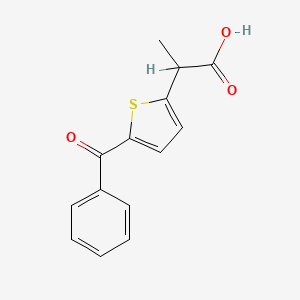 |
0.353 | ||
| ENC000597 | 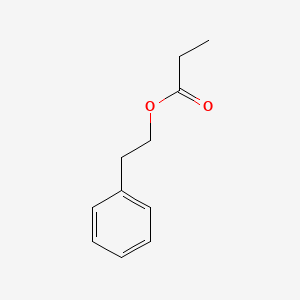 |
0.436 | D0G1VX |  |
0.348 | ||