NPs Basic Information
|
Name |
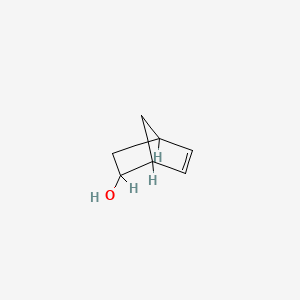
Bicyclo[2.2.1]hept-5-en-2-ol
|
| Molecular Formula | C7H10O | |
| IUPAC Name* |
bicyclo[2.2.1]hept-5-en-2-ol
|
|
| SMILES |
C1C2CC(C1C=C2)O
|
|
| InChI |
InChI=1S/C7H10O/c8-7-4-5-1-2-6(7)3-5/h1-2,5-8H,3-4H2
|
|
| InChIKey |
MKOSBHNWXFSHSW-UHFFFAOYSA-N
|
|
| Synonyms |
Bicyclo[2.2.1]hept-5-en-2-ol; 13080-90-5; 5-Norbornene-2-ol; 5-Norbornen-2-ol; 5-Norbornen-2-ol, exo-; exo-2-Norbornenol; Bicyclo(2.2.1)hept-5-en-2-ol; Bicyclo[2.2.1]hept-5-en-2-ol, exo-; bicyclo[2.2.1)hept-5-en-2-ol; Bicyclo(2.2.1)hept-5-en-2-ol, exo-; bicyclo[2.2.1]hept-2-en-5-ol; norbornene-5-ol; Norbornen-5-ol; 5-exo-norbornenol; 2890-98-4; endo-2-Norbornenol; NSC50234; EINECS 235-987-9; exo-Dehydronorborneol; endo-Dehydronorborneol; 5-Norbornen-2-ol, endo-; ghl.PD_Mitscher_leg0.740; SCHEMBL278517; DTXSID40871953; 5-bicyclo[2.2.1]hept-2-enol; ACT03083; MFCD00167566; NSC 50234; NSC-50234; NSC108290; NSC110578; AKOS015917582; Bicyclo[2.2.1]hept-5-en-2-ol #; CS-W008466; FD14040; NSC-108290; NSC-110578; DA-17372; DS-13830; Bicyclo[2.2.1]hept-5-en-2-ol, endo-; FT-0689543; EN300-110065; A806152; A905540; 694-97-3
|
|
| CAS | 13080-90-5 | |
| PubChem CID | 96066 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 110.15 | ALogp: | 0.8 |
| HBD: | 1 | HBA: | 1 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 20.2 | Aromatic Rings: | 2 |
| Heavy Atoms: | 8 | QED Weighted: | 0.467 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.417 | MDCK Permeability: | 0.00008530 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.005 |
| Human Intestinal Absorption (HIA): | 0.075 | 20% Bioavailability (F20%): | 0.007 |
| 30% Bioavailability (F30%): | 0.279 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.996 | Plasma Protein Binding (PPB): | 31.42% |
| Volume Distribution (VD): | 0.938 | Fu: | 54.84% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.129 | CYP1A2-substrate: | 0.85 |
| CYP2C19-inhibitor: | 0.038 | CYP2C19-substrate: | 0.67 |
| CYP2C9-inhibitor: | 0.019 | CYP2C9-substrate: | 0.482 |
| CYP2D6-inhibitor: | 0.012 | CYP2D6-substrate: | 0.709 |
| CYP3A4-inhibitor: | 0.125 | CYP3A4-substrate: | 0.238 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 13.758 | Half-life (T1/2): | 0.673 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.006 | Human Hepatotoxicity (H-HT): | 0.014 |
| Drug-inuced Liver Injury (DILI): | 0.032 | AMES Toxicity: | 0.048 |
| Rat Oral Acute Toxicity: | 0.305 | Maximum Recommended Daily Dose: | 0.056 |
| Skin Sensitization: | 0.103 | Carcinogencity: | 0.172 |
| Eye Corrosion: | 0.384 | Eye Irritation: | 0.978 |
| Respiratory Toxicity: | 0.144 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC002637 | 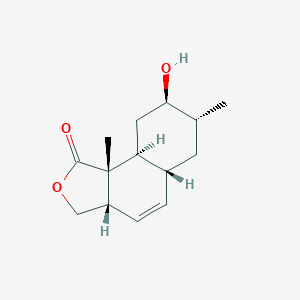 |
0.296 | D0B4TN | 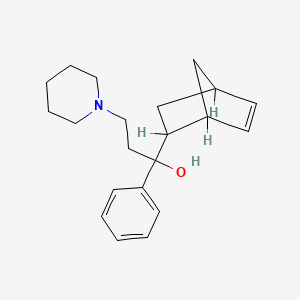 |
0.247 | ||
| ENC002165 | 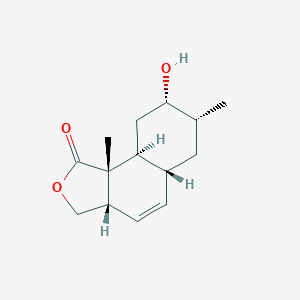 |
0.296 | D03CNS | 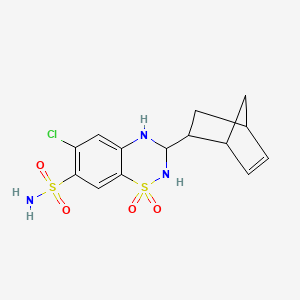 |
0.236 | ||
| ENC001275 |  |
0.289 | D04CSZ | 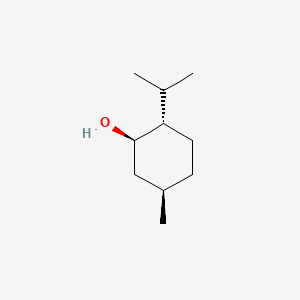 |
0.209 | ||
| ENC005380 | 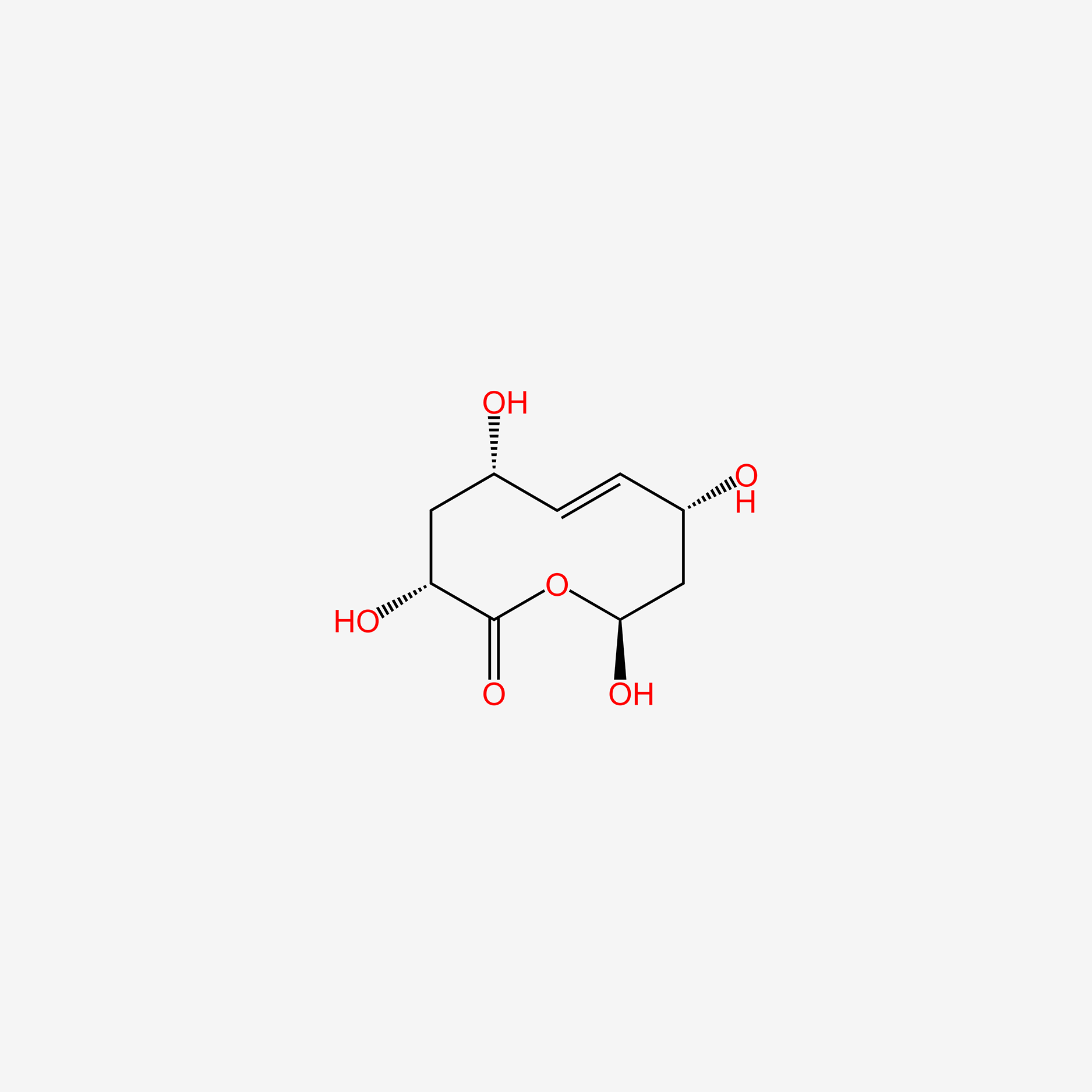 |
0.260 | D0Z4EI | 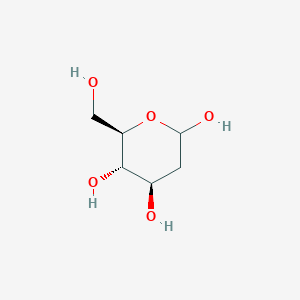 |
0.182 | ||
| ENC001252 | 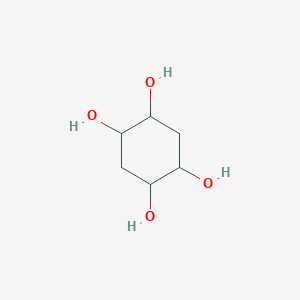 |
0.256 | D07HZY | 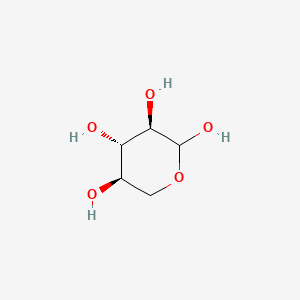 |
0.167 | ||
| ENC002508 | 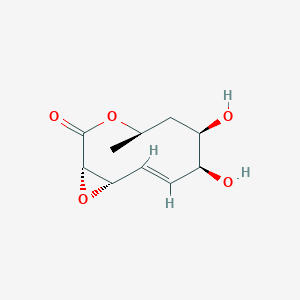 |
0.231 | D0T3AD | 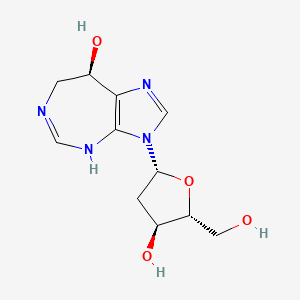 |
0.167 | ||
| ENC003771 | 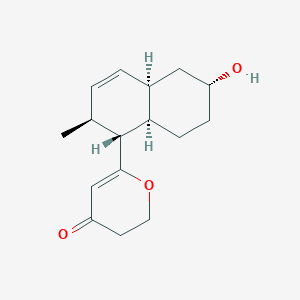 |
0.222 | D06VFO | 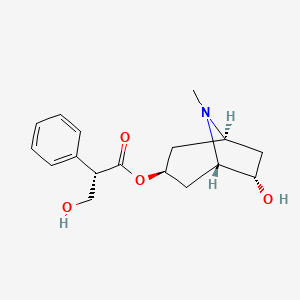 |
0.164 | ||
| ENC001433 | 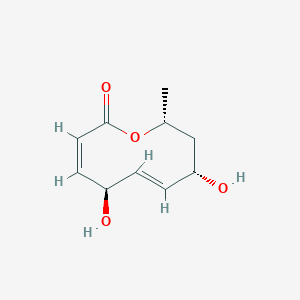 |
0.220 | D0YS7D | 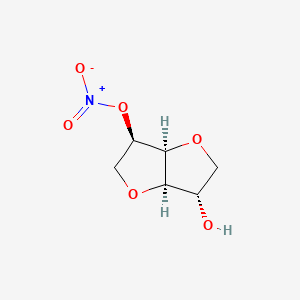 |
0.157 | ||
| ENC002454 | 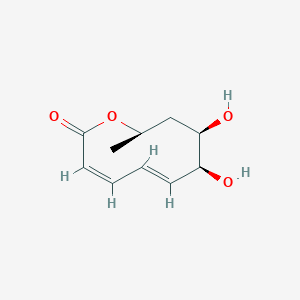 |
0.220 | D0MU9L | 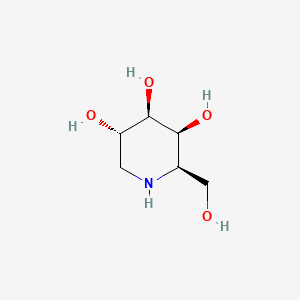 |
0.156 | ||
| ENC001184 | 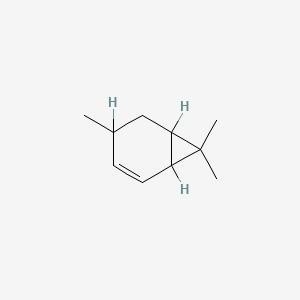 |
0.220 | D0X5XU | 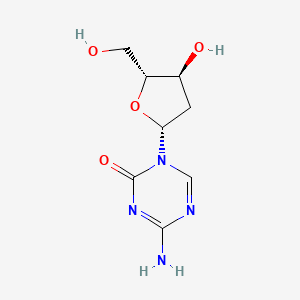 |
0.155 | ||