NPs Basic Information
|
Name |
Lycorine
|
| Molecular Formula | C16H17NO4 | |
| IUPAC Name* |
(1S,17S,18S,19S)-5,7-dioxa-12-azapentacyclo[10.6.1.02,10.04,8.015,19]nonadeca-2,4(8),9,15-tetraene-17,18-diol
|
|
| SMILES |
C1CN2CC3=CC4=C(C=C3[C@H]5[C@H]2C1=C[C@@H]([C@H]5O)O)OCO4
|
|
| InChI |
InChI=1S/C16H17NO4/c18-11-3-8-1-2-17-6-9-4-12-13(21-7-20-12)5-10(9)14(15(8)17)16(11)19/h3-5,11,14-16,18-19H,1-2,6-7H2/t11-,14-,15+,16+/m0/s1
|
|
| InChIKey |
XGVJWXAYKUHDOO-DANNLKNASA-N
|
|
| Synonyms |
lycorine; 476-28-8; Amarylline; Galanthidine; Narcissine; Licorine; (-)-Lycorine; NSC401360; NSC683873; CHEBI:6601; CHEMBL400092; I9Q105R5BU; GNF-PF-4974; Lycoran-1-alpha,2-beta-diol, 3,3-alpha-didehydro-; NSC-401360; NSC-683873; 9,10-(methylenedioxy)-3,12-didehydrogalanthan-1alpha,2beta-diol; NSC 401360; Galanthan-1,2-diol, 3,12-didehydro-9,10-(methylenebis(oxy))-, (1-alpha,2-beta)-; (1S,2S,3a1S,12bS)-2,3a1,4,5,7,12b-hexahydro-1H-[1,3]dioxolo[4,5-j]pyrrolo[3,2,1-de]phenanthridine-1,2-diol; (1S,2S,12bS,12cS)-2,4,5,7,12b,12c-hexahydro-1H-[1,3]dioxolo[4,5-j]pyrrolo[3,2,1-de]phenanthridine-1,2-diol; EINECS 207-503-6; UNII-I9Q105R5BU; 3,3a-Didehydrolycoran-1alpha,2beta-diol; Likorin; (1S,2S,12BS,12CS)-2,4,5,7,12B,12C-HEXAHYDRO-1H-(1,3)DIOXOLO(4,5-J)PYRROLO(3,2,1-DE)PHENANTHRIDINE-1,2-DIOL; 3,2.beta.-diol; Lycoran-1.alpha.,2.beta.-diol, 3,3a-didehydro-; LYCORINE [MI]; Lycoran-1alpha,2beta-diol, 3,3a-didehydro-; BSPBio_001302; KBioGR_000022; KBioSS_000022; SCHEMBL626071; BCBcMAP01_000100; KBio2_000022; KBio2_002590; KBio2_005158; KBio3_000043; KBio3_000044; DTXSID60197208; Bio2_000022; Bio2_000502; HMS1361B04; HMS1791B04; HMS1989B04; HMS3402B04; HMS3885I12; HY-N0288; ZINC3881372; BDBM50221066; MFCD00221746; NSC781764; s3903; Lycoran-1.alpha., 3,3a-didehydro-; AKOS000278045; CCG-208232; NSC-781764; IDI1_033772; QTL1_000052; SMP1_000184; NCGC00163413-01; NCGC00163413-02; NCGC00163413-03; NCGC00163413-07; NCI60_003767; CS-0008781; C08532; Q420314; BRD-K64909280-001-02-7; 3,12-[methylenebis(oxy)]-galanthan-1.alpha.,2.beta.-diol; Galanthan-1, 3,12-didehydro-9,10-[methylenebis(oxy)]-, (1.alpha.,2.beta.)-; Galanthan-1, 3,4-didehydro-11,12-[methylenebis(oxy)]-, (1.alpha.,2.beta.)-; (1S,17S,18S,19S)-5,7-dioxa-12-azapentacyclo[10.6.1.0?,??.0?,?.0??,??]nonadeca-2,4(8),9,15-tetraene-17,18-diol; (1S,17S,18S,19S)-5,7-dioxa-12-azapentacyclo[10.6.1.02,10.04,8.015,19]nonadeca-2,4(8),9,15-tetraene-17,18-diol; 1H-[1,3]Dioxolo[4,5-j]pyrrolo[3,2,1-de]phenanthridine-1,2-diol, 2,4,5,7,12b,12c-hexahydro-, (1S,2S,12bS,12cS)-; 2,4,5,7,12b,12c-Hexahydro-1H-[1,3]dioxolo[4,5-j]pyrrolo[3,2,1-de]phenanthridine-1,2-diol, (1S,2S,12bS, 12cS)-; 2,5,7,12b,12c-Hexahydro-1H-[1,3]-dioxolo[4,5-j]pyrrolo[3,2,1-de]phenanthridine-1,2-diol; 3KD
|
|
| CAS | 476-28-8 | |
| PubChem CID | 72378 | |
| ChEMBL ID | CHEMBL400092 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 287.31 | ALogp: | 0.0 |
| HBD: | 2 | HBA: | 5 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 62.2 | Aromatic Rings: | 5 |
| Heavy Atoms: | 21 | QED Weighted: | 0.701 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.654 | MDCK Permeability: | 0.00003040 |
| Pgp-inhibitor: | 0.004 | Pgp-substrate: | 0.499 |
| Human Intestinal Absorption (HIA): | 0.005 | 20% Bioavailability (F20%): | 0.129 |
| 30% Bioavailability (F30%): | 0.19 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.983 | Plasma Protein Binding (PPB): | 71.25% |
| Volume Distribution (VD): | 2.527 | Fu: | 25.39% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.712 | CYP1A2-substrate: | 0.659 |
| CYP2C19-inhibitor: | 0.095 | CYP2C19-substrate: | 0.81 |
| CYP2C9-inhibitor: | 0.019 | CYP2C9-substrate: | 0.342 |
| CYP2D6-inhibitor: | 0.938 | CYP2D6-substrate: | 0.733 |
| CYP3A4-inhibitor: | 0.152 | CYP3A4-substrate: | 0.32 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 16.253 | Half-life (T1/2): | 0.479 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.023 | Human Hepatotoxicity (H-HT): | 0.057 |
| Drug-inuced Liver Injury (DILI): | 0.106 | AMES Toxicity: | 0.124 |
| Rat Oral Acute Toxicity: | 0.859 | Maximum Recommended Daily Dose: | 0.929 |
| Skin Sensitization: | 0.05 | Carcinogencity: | 0.945 |
| Eye Corrosion: | 0.003 | Eye Irritation: | 0.013 |
| Respiratory Toxicity: | 0.965 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
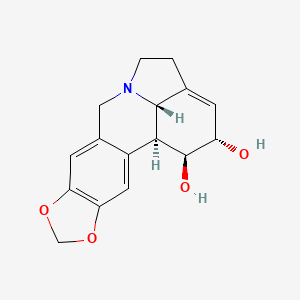
| ENC001089 | 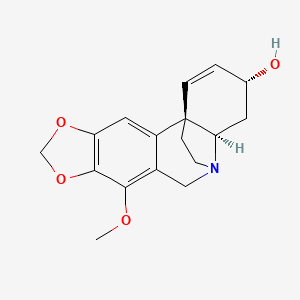 |
0.302 | D0L1JW |  |
0.330 | ||
| ENC001059 | 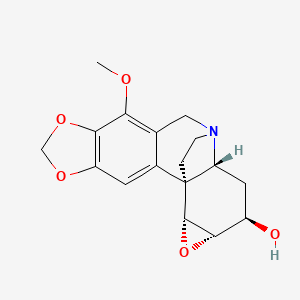 |
0.293 | D0D4HN | 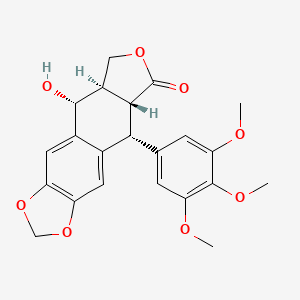 |
0.292 | ||
| ENC000361 | 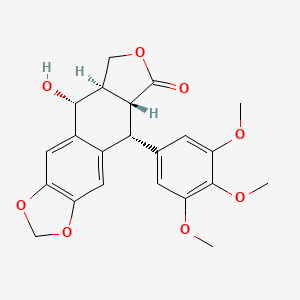 |
0.292 | D0M4XY | 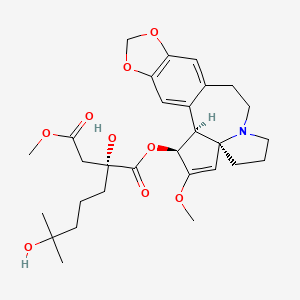 |
0.278 | ||
| ENC001035 | 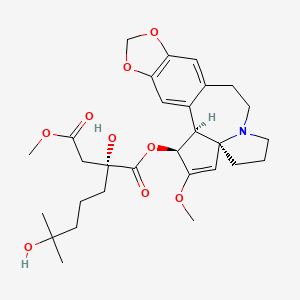 |
0.278 | D07UXP | 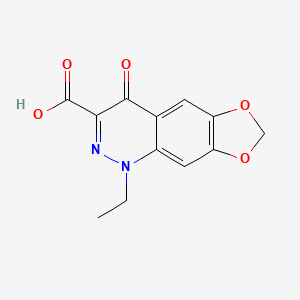 |
0.267 | ||
| ENC005030 | 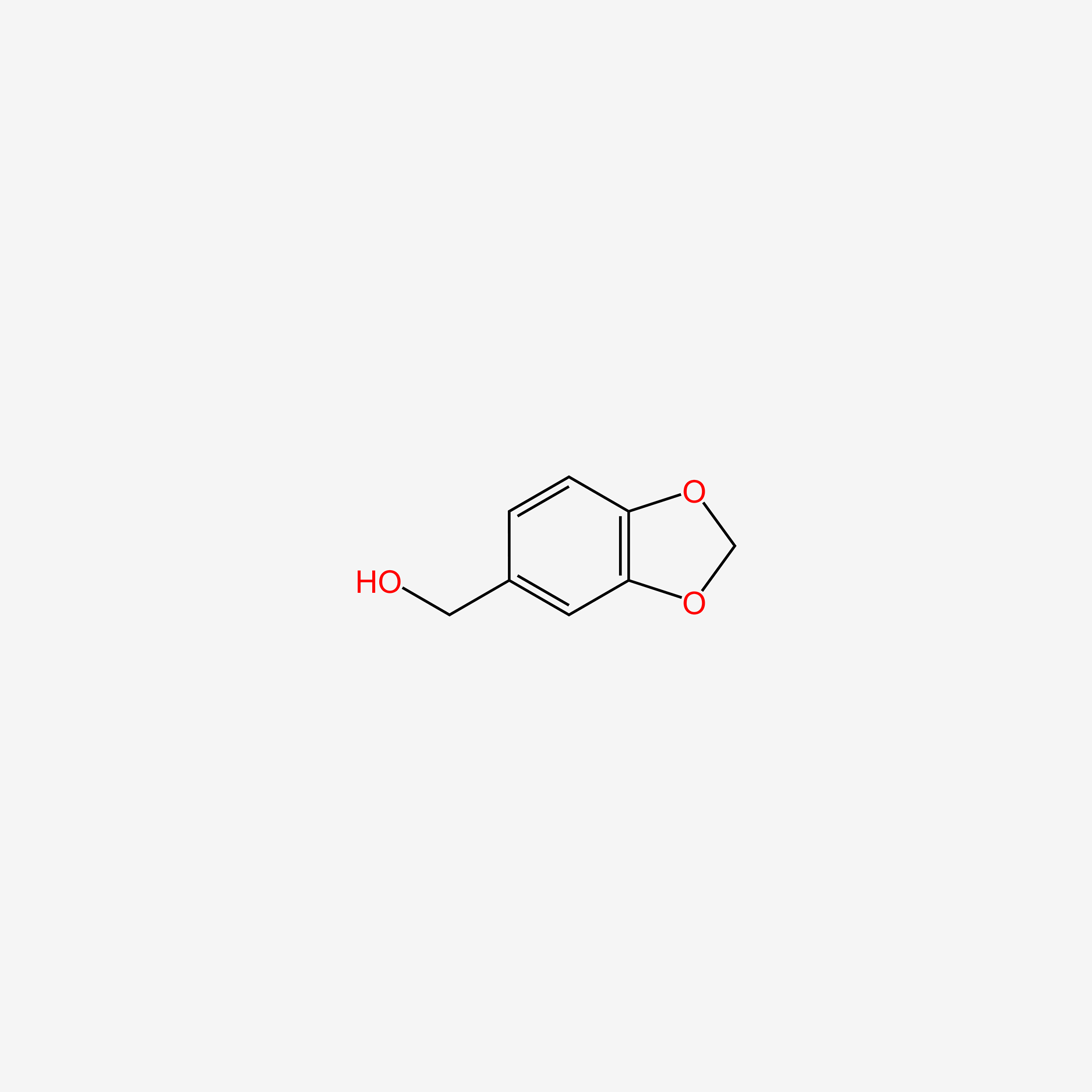 |
0.274 | D0W8WB | 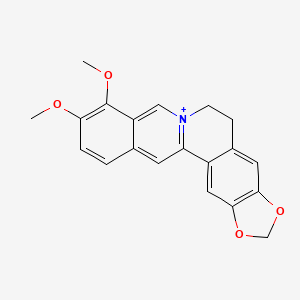 |
0.264 | ||
| ENC001881 |  |
0.266 | D09NIB | 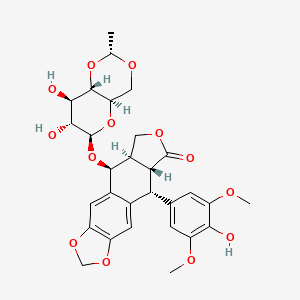 |
0.259 | ||
| ENC000812 | 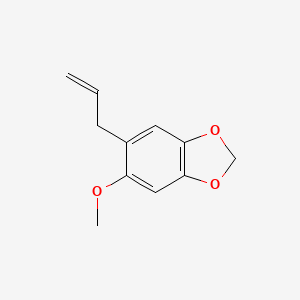 |
0.263 | D04TDQ | 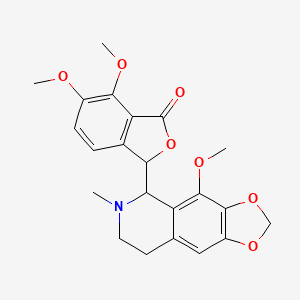 |
0.248 | ||
| ENC003417 | 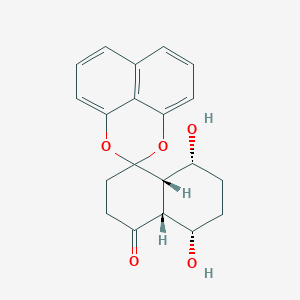 |
0.243 | D06GDY | 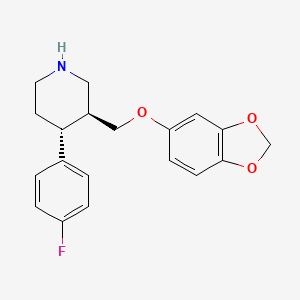 |
0.245 | ||
| ENC001426 | 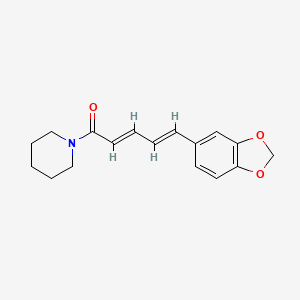 |
0.242 | D01DBQ |  |
0.237 | ||
| ENC001086 |  |
0.224 | D05MQK |  |
0.229 | ||