NPs Basic Information
|
Name |
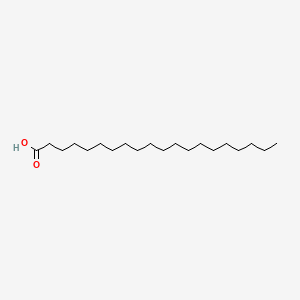
Arachidic Acid
|
| Molecular Formula | C20H40O2 | |
| IUPAC Name* |
icosanoic acid
|
|
| SMILES |
CCCCCCCCCCCCCCCCCCCC(=O)O
|
|
| InChI |
InChI=1S/C20H40O2/c1-2-3-4-5-6-7-8-9-10-11-12-13-14-15-16-17-18-19-20(21)22/h2-19H2,1H3,(H,21,22)
|
|
| InChIKey |
VKOBVWXKNCXXDE-UHFFFAOYSA-N
|
|
| Synonyms |
Arachidic acid; Icosanoic acid; EICOSANOIC ACID; 506-30-9; Arachic acid; n-Eicosanoic acid; Arachidinic acid; arachidate; eicosanoate; Elcosanoic Acid; CHEBI:28822; C20:0; PQB8MJD4RB; n-eicosanoate; NSC-93983; Eicosanoicacid; NSC 93983; UNII-PQB8MJD4RB; EINECS 208-031-3; MFCD00002755; Arachinsaeure; Icosanoicacid; Arachate; eicosatrienoic-acid; EICOSANO1C ACID; Arachidic acid (synthetic); Arachidic acid, >=99%; EC 208-031-3; SCHEMBL6539; ARACHIDIC ACID [MI]; WLN: QV19; ARACHIDIC ACID [INCI]; CH3-[CH2]18-COOH; CHEMBL1173381; DTXSID1060134; FA 20:0a; Arachidic acid, analytical standard; NSC93983; ZINC6920376; BBL027402; BDBM50463966; LMFA01010020; s5570; STL373057; AKOS015839825; CCG-267612; CS-W004260; HY-W004260; AC-15197; AS-14639; DB-051805; AM20100241; Arachidic acid, synthetic, >=99.0% (GC); FT-0625651; FT-0625652; Arachidic acid, Vetec(TM) reagent grade, 99%; C06425; O11825; A828210; Q409608; 38660976-A573-4A36-B283-4EA09D1E22EC; icosanoic acid, eicosanoic acid, n-eicosanoic acid, arachic acid,
|
|
| CAS | 506-30-9 | |
| PubChem CID | 10467 | |
| ChEMBL ID | CHEMBL1173381 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 312.5 | ALogp: | 8.5 |
| HBD: | 1 | HBA: | 2 |
| Rotatable Bonds: | 18 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 37.3 | Aromatic Rings: | 0 |
| Heavy Atoms: | 22 | QED Weighted: | 0.275 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.099 | MDCK Permeability: | 0.00001740 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0 |
| Human Intestinal Absorption (HIA): | 0.005 | 20% Bioavailability (F20%): | 0.427 |
| 30% Bioavailability (F30%): | 0.996 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.017 | Plasma Protein Binding (PPB): | 99.13% |
| Volume Distribution (VD): | 0.887 | Fu: | 0.82% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.209 | CYP1A2-substrate: | 0.174 |
| CYP2C19-inhibitor: | 0.265 | CYP2C19-substrate: | 0.062 |
| CYP2C9-inhibitor: | 0.092 | CYP2C9-substrate: | 0.993 |
| CYP2D6-inhibitor: | 0.014 | CYP2D6-substrate: | 0.036 |
| CYP3A4-inhibitor: | 0.05 | CYP3A4-substrate: | 0.011 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 2.523 | Half-life (T1/2): | 0.356 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.102 | Human Hepatotoxicity (H-HT): | 0.02 |
| Drug-inuced Liver Injury (DILI): | 0.048 | AMES Toxicity: | 0.005 |
| Rat Oral Acute Toxicity: | 0.019 | Maximum Recommended Daily Dose: | 0.014 |
| Skin Sensitization: | 0.942 | Carcinogencity: | 0.041 |
| Eye Corrosion: | 0.975 | Eye Irritation: | 0.95 |
| Respiratory Toxicity: | 0.891 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000282 |  |
0.913 | D00AOJ |  |
0.711 | ||
| ENC000110 |  |
0.905 | D07ILQ |  |
0.662 | ||
| ENC000356 | 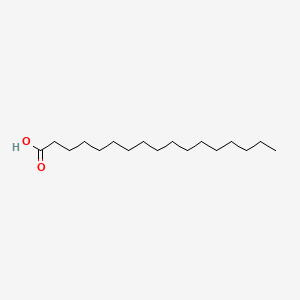 |
0.857 | D0O1PH |  |
0.573 | ||
| ENC000474 | 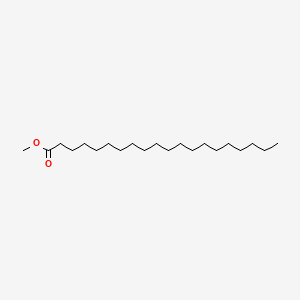 |
0.817 | D00FGR | 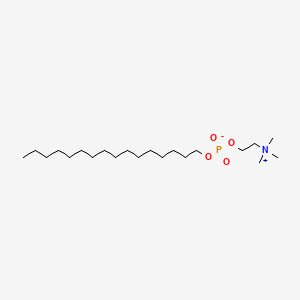 |
0.516 | ||
| ENC000050 | 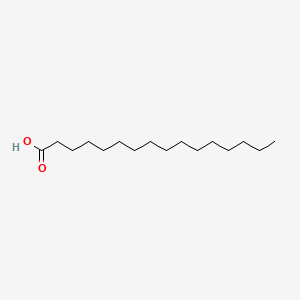 |
0.810 | D0Z5SM |  |
0.500 | ||
| ENC000358 |  |
0.808 | D00STJ | 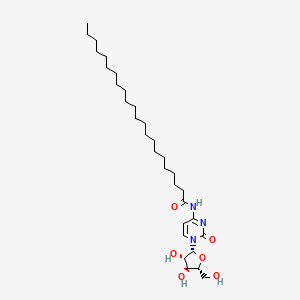 |
0.500 | ||
| ENC000745 | 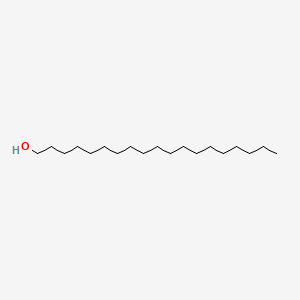 |
0.806 | D05ATI |  |
0.429 | ||
| ENC000285 | 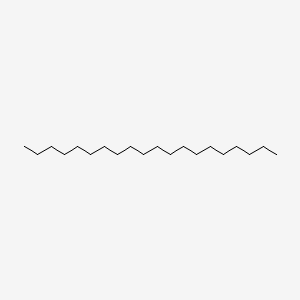 |
0.779 | D0XN8C | 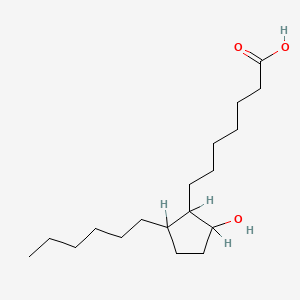 |
0.419 | ||
| ENC000497 |  |
0.775 | D0P1RL |  |
0.396 | ||
| ENC000431 |  |
0.771 | D0Z5BC |  |
0.394 | ||