NPs Basic Information
|
Name |
12-Methyltetradecanoic acid
|
| Molecular Formula | C15H30O2 | |
| IUPAC Name* |
12-methyltetradecanoic acid
|
|
| SMILES |
CCC(C)CCCCCCCCCCC(=O)O
|
|
| InChI |
InChI=1S/C15H30O2/c1-3-14(2)12-10-8-6-4-5-7-9-11-13-15(16)17/h14H,3-13H2,1-2H3,(H,16,17)
|
|
| InChIKey |
XKLJLHAPJBUBNL-UHFFFAOYSA-N
|
|
| Synonyms |
12-Methyltetradecanoic acid; 5502-94-3; Sarcinic acid; Aseanostatin P5; ANTEISOPENTADECANOIC ACID; 12-methyl myristic acid; 12-methyl-tetradecanoic acid; methyl myristic acid; 12-MTA; anteiso-C15:0; Tetradecanoic acid, 12-methyl-; TP0OL0Z8US; CHEBI:39251; 12-METHYLTETRADECANOICACID; anteiso-15:0; UNII-TP0OL0Z8US; 12-Methyl tetradecanoic acid; anteiso-C15; 15:0 anteiso; 12-methylmyristic acid; 12-Methyltetradecansaeure; 12-Methyl-tetradecansaeure; C15:0ai; AI-PENTADECANOIC ACID; 15:0ai; MLS000517264; SCHEMBL397325; (+)-12-methyl myristic acid; CHEMBL495852; aC15:0; DTXSID10970381; (+)-12-Methyltetradecanoic acid; HMS2267L13; METHYL MYRISTIC ACID [INCI]; a15:0; LMFA01020008; NCGC00247048-01; SMR000127417; (+/-)-12-METHYLTETRADECANOIC ACID; FT-0769402; Q27119789; 12-Methyltetradecanoic acid; Sarcinic acid; Aseanostatin P5
|
|
| CAS | 5502-94-3 | |
| PubChem CID | 21672 | |
| ChEMBL ID | CHEMBL495852 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 242.4 | ALogp: | 5.5 |
| HBD: | 1 | HBA: | 2 |
| Rotatable Bonds: | 12 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 37.3 | Aromatic Rings: | 0 |
| Heavy Atoms: | 17 | QED Weighted: | 0.468 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.782 | MDCK Permeability: | 0.00002340 |
| Pgp-inhibitor: | 0.048 | Pgp-substrate: | 0 |
| Human Intestinal Absorption (HIA): | 0.004 | 20% Bioavailability (F20%): | 0.708 |
| 30% Bioavailability (F30%): | 0.967 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.138 | Plasma Protein Binding (PPB): | 98.18% |
| Volume Distribution (VD): | 0.407 | Fu: | 1.17% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.195 | CYP1A2-substrate: | 0.23 |
| CYP2C19-inhibitor: | 0.074 | CYP2C19-substrate: | 0.301 |
| CYP2C9-inhibitor: | 0.358 | CYP2C9-substrate: | 0.984 |
| CYP2D6-inhibitor: | 0.006 | CYP2D6-substrate: | 0.055 |
| CYP3A4-inhibitor: | 0.019 | CYP3A4-substrate: | 0.031 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 2.488 | Half-life (T1/2): | 0.673 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.021 | Human Hepatotoxicity (H-HT): | 0.034 |
| Drug-inuced Liver Injury (DILI): | 0.039 | AMES Toxicity: | 0.005 |
| Rat Oral Acute Toxicity: | 0.021 | Maximum Recommended Daily Dose: | 0.025 |
| Skin Sensitization: | 0.802 | Carcinogencity: | 0.07 |
| Eye Corrosion: | 0.972 | Eye Irritation: | 0.972 |
| Respiratory Toxicity: | 0.875 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
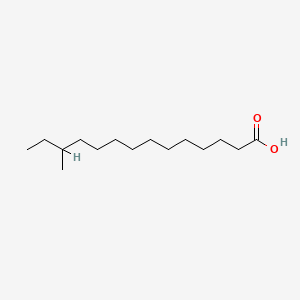
| ENC000916 | 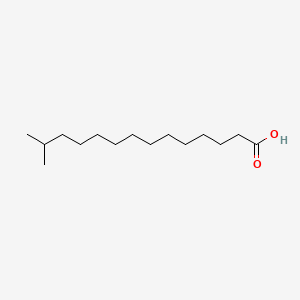 |
0.774 | D0Z5BC | 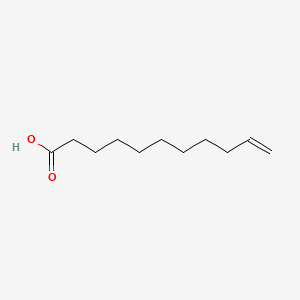 |
0.509 | ||
| ENC000549 |  |
0.764 | D0O1PH |  |
0.487 | ||
| ENC001913 | 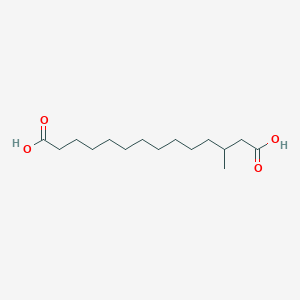 |
0.737 | D0P1RL | 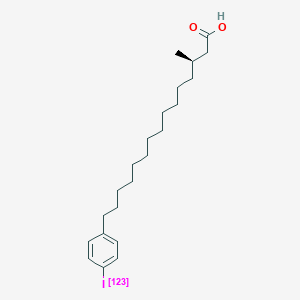 |
0.475 | ||
| ENC000102 | 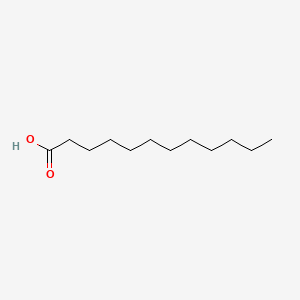 |
0.720 | D0XN8C | 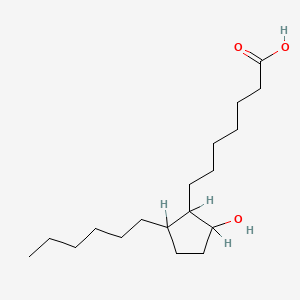 |
0.452 | ||
| ENC001142 | 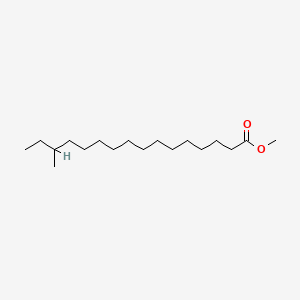 |
0.689 | D07ILQ |  |
0.446 | ||
| ENC001596 | 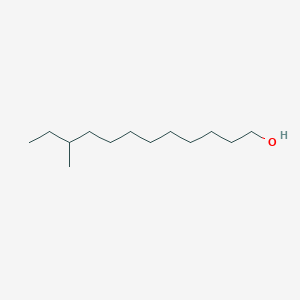 |
0.686 | D0E4WR | 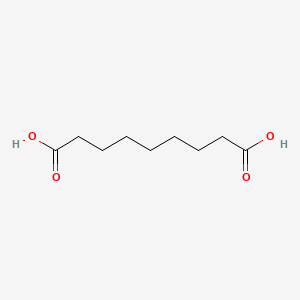 |
0.439 | ||
| ENC000378 | 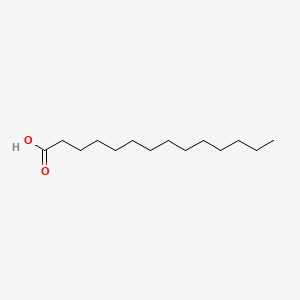 |
0.673 | D05ATI |  |
0.424 | ||
| ENC000270 | 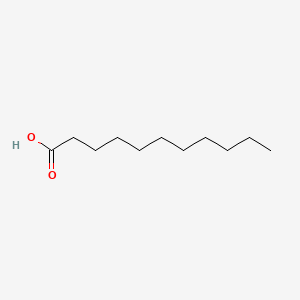 |
0.660 | D0I4DQ |  |
0.398 | ||
| ENC001228 |  |
0.654 | D0G2KD |  |
0.397 | ||
| ENC000850 |  |
0.654 | D0O1TC |  |
0.392 | ||