NPs Basic Information
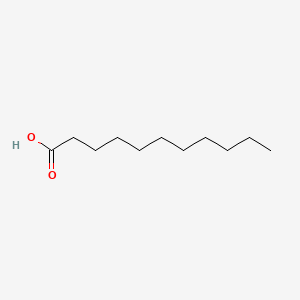
|
Name |
Undecanoic acid
|
| Molecular Formula | C11H22O2 | |
| IUPAC Name* |
undecanoic acid
|
|
| SMILES |
CCCCCCCCCCC(=O)O
|
|
| InChI |
InChI=1S/C11H22O2/c1-2-3-4-5-6-7-8-9-10-11(12)13/h2-10H2,1H3,(H,12,13)
|
|
| InChIKey |
ZDPHROOEEOARMN-UHFFFAOYSA-N
|
|
| Synonyms |
UNDECANOIC ACID; 112-37-8; Hendecanoic acid; Undecylic acid; undecanoate; n-Undecanoic acid; n-Undecoic acid; n-Undecylic acid; Undecoic acid; 1-Decanecarboxylic acid; FEMA No. 3245; undecanoicacid; Undecanoate [USAN]; Undekansaeure; NSC-7885; CHEBI:32368; MFCD00002730; Undecanoate;Hendecanoic acid; 138ON3IIQG; NSC7885; 1-decanecarboxylate; C11:0; NSC 7885; EINECS 203-964-2; UNII-138ON3IIQG; BRN 1759287; AI3-02280; undecansäure; Undecanoic acid, 98%; DSSTox_CID_1690; bmse000563; SCHEMBL9266; DSSTox_RID_76286; NCIOpen2_009435; DSSTox_GSID_21690; 4-02-00-01068 (Beilstein Handbook Reference); WLN: QV10; CH3-[CH2]9-COOH; UNDECANOIC ACID [FHFI]; UNDECANOIC ACID [INCI]; CHEMBL108030; GTPL5533; QSPL 035; QSPL 155; QSPL 192; DTXSID8021690; Undecanoic acid, >=97%, FG; HMS3740G13; ZINC1586297; Tox21_201031; Undecanoic acid, analytical standard; BDBM50511006; LMFA01010011; AKOS000276906; AM85321; CS-W004282; DS-6048; HY-W004282; PB44018; NCGC00248901-01; NCGC00258584-01; BP-27912; CAS-112-37-8; Hendecanoic Acid/Undecanoic Acid(C11:0); SY007756; FT-0689759; S9454; U0004; EN300-19485; C17715; P19940; A802562; Q425988; J-002761; 75EF58E9-1C6B-4875-8B82-D0B7A10FAEA2; F2191-0248; Z104473988
|
|
| CAS | 112-37-8 | |
| PubChem CID | 8180 | |
| ChEMBL ID | CHEMBL108030 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 186.29 | ALogp: | 3.7 |
| HBD: | 1 | HBA: | 2 |
| Rotatable Bonds: | 9 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 37.3 | Aromatic Rings: | 0 |
| Heavy Atoms: | 13 | QED Weighted: | 0.544 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.871 | MDCK Permeability: | 0.00003170 |
| Pgp-inhibitor: | 0.065 | Pgp-substrate: | 0 |
| Human Intestinal Absorption (HIA): | 0.005 | 20% Bioavailability (F20%): | 0.865 |
| 30% Bioavailability (F30%): | 0.973 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.66 | Plasma Protein Binding (PPB): | 95.57% |
| Volume Distribution (VD): | 0.287 | Fu: | 2.82% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.111 | CYP1A2-substrate: | 0.314 |
| CYP2C19-inhibitor: | 0.033 | CYP2C19-substrate: | 0.515 |
| CYP2C9-inhibitor: | 0.188 | CYP2C9-substrate: | 0.979 |
| CYP2D6-inhibitor: | 0.007 | CYP2D6-substrate: | 0.103 |
| CYP3A4-inhibitor: | 0.01 | CYP3A4-substrate: | 0.033 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 2.361 | Half-life (T1/2): | 0.786 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.024 | Human Hepatotoxicity (H-HT): | 0.036 |
| Drug-inuced Liver Injury (DILI): | 0.043 | AMES Toxicity: | 0.006 |
| Rat Oral Acute Toxicity: | 0.059 | Maximum Recommended Daily Dose: | 0.017 |
| Skin Sensitization: | 0.549 | Carcinogencity: | 0.109 |
| Eye Corrosion: | 0.98 | Eye Irritation: | 0.987 |
| Respiratory Toxicity: | 0.649 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000102 | 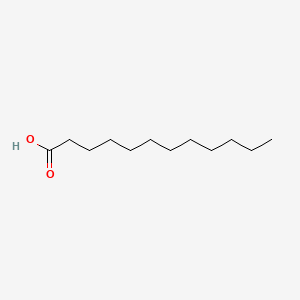 |
0.923 | D0Z5BC | 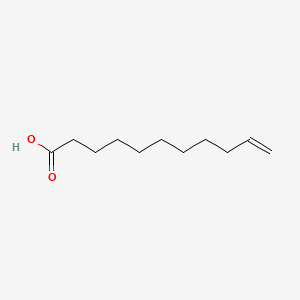 |
0.636 | ||
| ENC000088 | 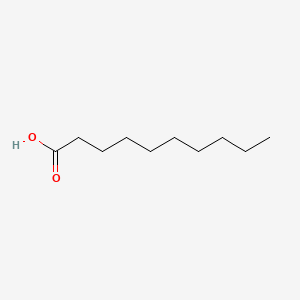 |
0.917 | D0XN8C | 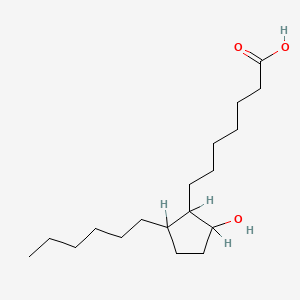 |
0.557 | ||
| ENC000263 | 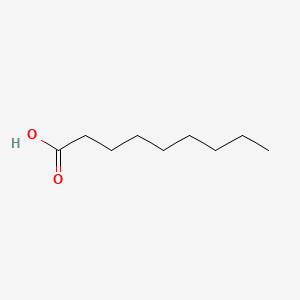 |
0.833 | D0O1PH |  |
0.545 | ||
| ENC000378 | 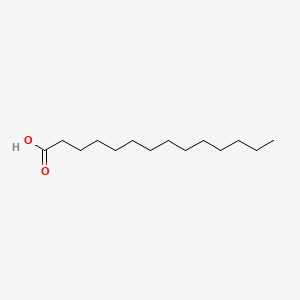 |
0.800 | D0E4WR | 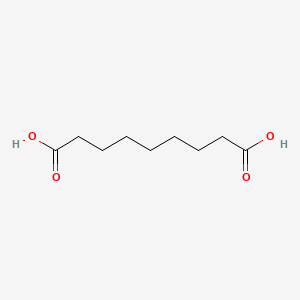 |
0.543 | ||
| ENC000556 | 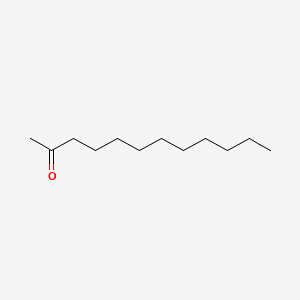 |
0.756 | D07ILQ |  |
0.500 | ||
| ENC000030 | 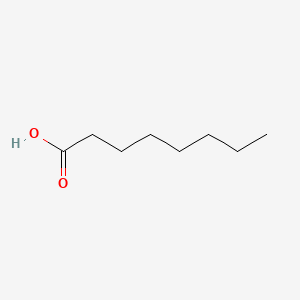 |
0.750 | D05ATI |  |
0.482 | ||
| ENC000466 | 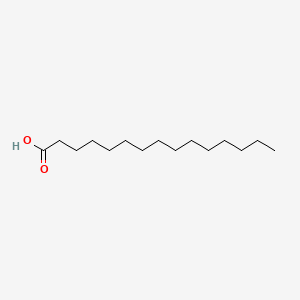 |
0.750 | D0O1TC | 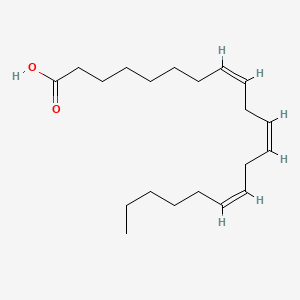 |
0.478 | ||
| ENC000050 | 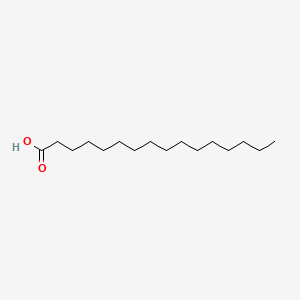 |
0.706 | D0FD0H | 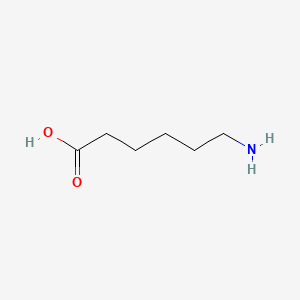 |
0.463 | ||
| ENC000472 | 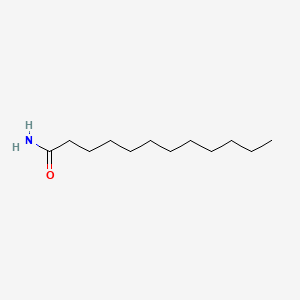 |
0.705 | D03ZJE | 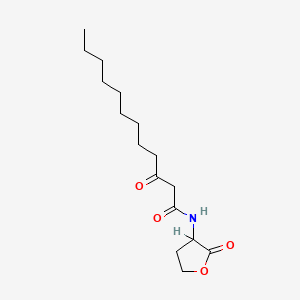 |
0.439 | ||
| ENC000399 |  |
0.705 | D0I4DQ |  |
0.438 | ||