NPs Basic Information
|
Name |
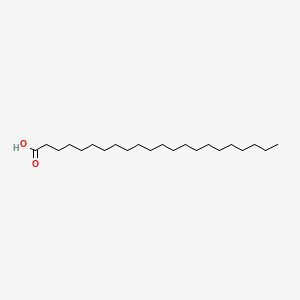
Docosanoic acid
|
| Molecular Formula | C22H44O2 | |
| IUPAC Name* |
docosanoic acid
|
|
| SMILES |
CCCCCCCCCCCCCCCCCCCCCC(=O)O
|
|
| InChI |
InChI=1S/C22H44O2/c1-2-3-4-5-6-7-8-9-10-11-12-13-14-15-16-17-18-19-20-21-22(23)24/h2-21H2,1H3,(H,23,24)
|
|
| InChIKey |
UKMSUNONTOPOIO-UHFFFAOYSA-N
|
|
| Synonyms |
Docosanoic acid; Behenic acid; 112-85-6; 1-Docosanoic acid; N-DOCOSANOIC ACID; Hydrofol Acid 560; Hydrofol 2022-55; Glycon B-70; Docosoic acid; Hystrene 5522; Hystrene 9022; Glycon B 70; CHEBI:28941; Prifrac 2989; Edenor C 22-85R; EXL 5; C22:0; MFCD00002807; NSC-32364; H390488X0A; B 95; Docosanoic acid (Chunks or pellets or flakes); HSDB 5578; EINECS 204-010-8; NSC 32364; Behensaeure; Docosansaeure; Dokosansaeure; Docosanic acid; n-Docosanoate; AI3-52709; 1-Docosanoate; UNII-H390488X0A; docosanoyl alcohol; Behenic Acid 85%; Behenic acid, 99%; CRODACID B; ORISTAR BA; Prifac 2987; Behenic Acid, Technical; BEHENIC ACID [MI]; EC 204-010-8; SCHEMBL6579; BEHENIC ACID [INCI]; PRIFRAC 2987; NAA 22S; Behenic acid; Docosanoic acid; DOCOSANOIC ACID [HSDB]; NAA 222S; CH3-[CH2]20-COOH; CHEMBL1173474; DTXSID3026930; AGP-103; Behenic acid, analytical standard; NSC32364; ZINC6920378; BBL025601; BDBM50488776; LMFA01010022; s5381; STL146320; AKOS005720830; CCG-267927; CS-W013765; FA 22:0; HY-W013049; AS-54401; B1248; B1747; D0963; FT-0745232; C08281; P50011; A854667; Q422590; W-108636; E2AAC59F-4B8D-460C-9C6E-E4E82C905122; 08O
|
|
| CAS | 112-85-6 | |
| PubChem CID | 8215 | |
| ChEMBL ID | CHEMBL1173474 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 340.6 | ALogp: | 9.6 |
| HBD: | 1 | HBA: | 2 |
| Rotatable Bonds: | 20 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 37.3 | Aromatic Rings: | 0 |
| Heavy Atoms: | 24 | QED Weighted: | 0.22 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.14 | MDCK Permeability: | 0.00001370 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0 |
| Human Intestinal Absorption (HIA): | 0.005 | 20% Bioavailability (F20%): | 0.331 |
| 30% Bioavailability (F30%): | 0.998 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.01 | Plasma Protein Binding (PPB): | 98.97% |
| Volume Distribution (VD): | 1.13 | Fu: | 0.75% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.141 | CYP1A2-substrate: | 0.165 |
| CYP2C19-inhibitor: | 0.242 | CYP2C19-substrate: | 0.058 |
| CYP2C9-inhibitor: | 0.069 | CYP2C9-substrate: | 0.994 |
| CYP2D6-inhibitor: | 0.02 | CYP2D6-substrate: | 0.031 |
| CYP3A4-inhibitor: | 0.062 | CYP3A4-substrate: | 0.009 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 2.648 | Half-life (T1/2): | 0.272 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.14 | Human Hepatotoxicity (H-HT): | 0.018 |
| Drug-inuced Liver Injury (DILI): | 0.054 | AMES Toxicity: | 0.005 |
| Rat Oral Acute Toxicity: | 0.016 | Maximum Recommended Daily Dose: | 0.013 |
| Skin Sensitization: | 0.95 | Carcinogencity: | 0.035 |
| Eye Corrosion: | 0.973 | Eye Irritation: | 0.933 |
| Respiratory Toxicity: | 0.877 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000357 |  |
0.913 | D00AOJ |  |
0.789 | ||
| ENC000358 |  |
0.885 | D07ILQ |  |
0.613 | ||
| ENC000464 |  |
0.831 | D00STJ | 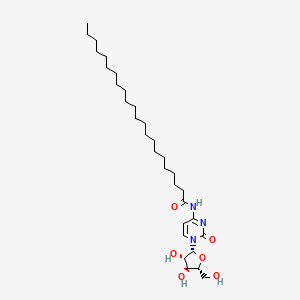 |
0.551 | ||
| ENC000110 | 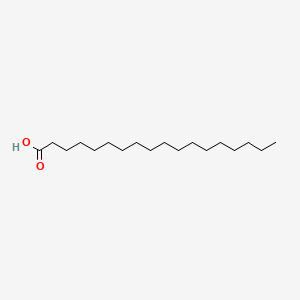 |
0.826 | D0O1PH |  |
0.534 | ||
| ENC000761 | 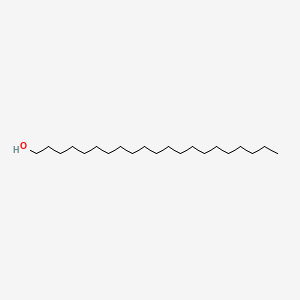 |
0.822 | D00FGR | 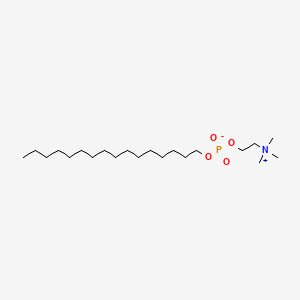 |
0.485 | ||
| ENC000432 |  |
0.797 | D0Z5SM |  |
0.464 | ||
| ENC000449 |  |
0.789 | D05ATI |  |
0.398 | ||
| ENC000356 |  |
0.783 | D0XN8C |  |
0.391 | ||
| ENC000431 |  |
0.781 | D0P1RL |  |
0.373 | ||
| ENC000724 |  |
0.771 | D0Z5BC |  |
0.364 | ||