NPs Basic Information
|
Name |
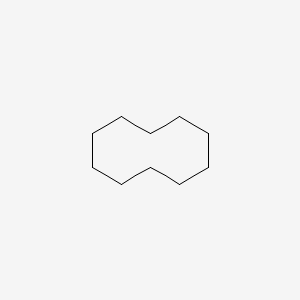
Cyclodecane
|
| Molecular Formula | C10H20 | |
| IUPAC Name* |
cyclodecane
|
|
| SMILES |
C1CCCCCCCCC1
|
|
| InChI |
InChI=1S/C10H20/c1-2-4-6-8-10-9-7-5-3-1/h1-10H2
|
|
| InChIKey |
LMGZGXSXHCMSAA-UHFFFAOYSA-N
|
|
| Synonyms |
CYCLODECANE; 293-96-9; 9N3JJ4GTR5; EINECS 206-032-3; cyclodecan; Cyclodecane, >=90%; UNII-9N3JJ4GTR5; DTXSID7059779; MFCD00003711; ZINC71613926; AKOS015915993; DB-047574; FT-0624160; D89414; A820228; Q118815; J-017495
|
|
| CAS | 293-96-9 | |
| PubChem CID | 9267 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 140.27 | ALogp: | 5.6 |
| HBD: | 0 | HBA: | 0 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 0.0 | Aromatic Rings: | 1 |
| Heavy Atoms: | 10 | QED Weighted: | 0.463 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.684 | MDCK Permeability: | 0.00001870 |
| Pgp-inhibitor: | 0.004 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.003 | 20% Bioavailability (F20%): | 0.339 |
| 30% Bioavailability (F30%): | 0.988 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.863 | Plasma Protein Binding (PPB): | 96.61% |
| Volume Distribution (VD): | 2.821 | Fu: | 2.90% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.896 | CYP1A2-substrate: | 0.22 |
| CYP2C19-inhibitor: | 0.561 | CYP2C19-substrate: | 0.09 |
| CYP2C9-inhibitor: | 0.238 | CYP2C9-substrate: | 0.931 |
| CYP2D6-inhibitor: | 0.064 | CYP2D6-substrate: | 0.143 |
| CYP3A4-inhibitor: | 0.094 | CYP3A4-substrate: | 0.072 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 4.086 | Half-life (T1/2): | 0.179 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.114 | Human Hepatotoxicity (H-HT): | 0.024 |
| Drug-inuced Liver Injury (DILI): | 0.304 | AMES Toxicity: | 0.028 |
| Rat Oral Acute Toxicity: | 0.051 | Maximum Recommended Daily Dose: | 0.041 |
| Skin Sensitization: | 0.948 | Carcinogencity: | 0.098 |
| Eye Corrosion: | 0.992 | Eye Irritation: | 0.98 |
| Respiratory Toxicity: | 0.889 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000324 |  |
0.833 | D00SBN |  |
0.286 | ||
| ENC000893 |  |
0.769 | D0N3PE | 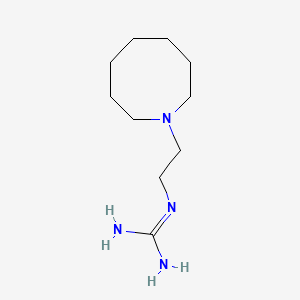 |
0.273 | ||
| ENC000840 | 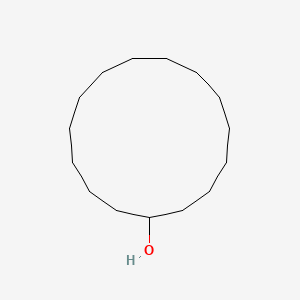 |
0.638 | D08VSI | 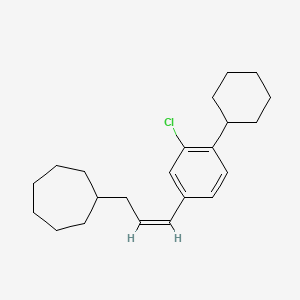 |
0.256 | ||
| ENC001017 | 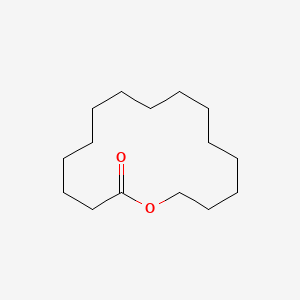 |
0.600 | D0S5NG | 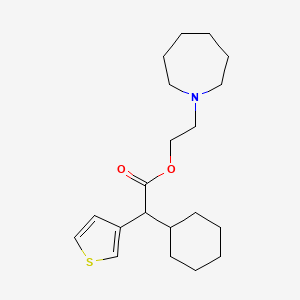 |
0.247 | ||
| ENC000251 | 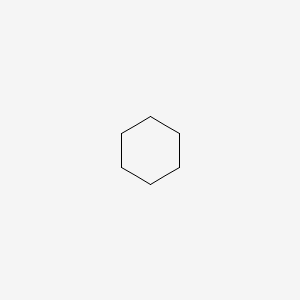 |
0.600 | D07XJM | 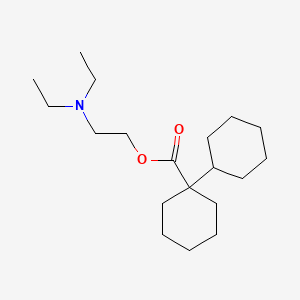 |
0.240 | ||
| ENC001146 | 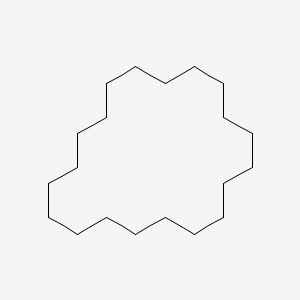 |
0.500 | D09GFL | 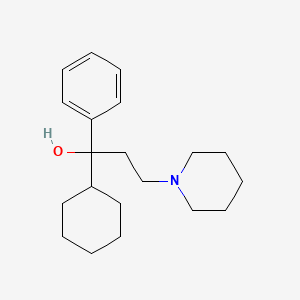 |
0.234 | ||
| ENC001147 | 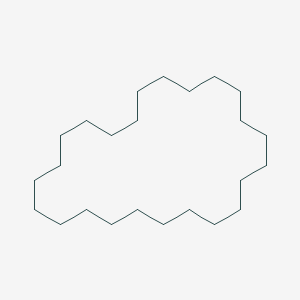 |
0.417 | D0L0MK | 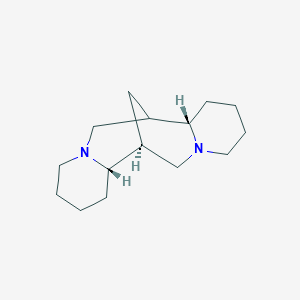 |
0.209 | ||
| ENC000170 | 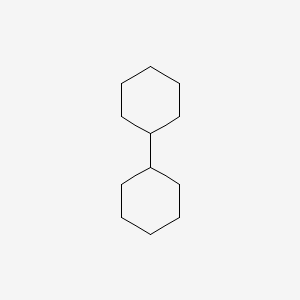 |
0.375 | D0R1WR | 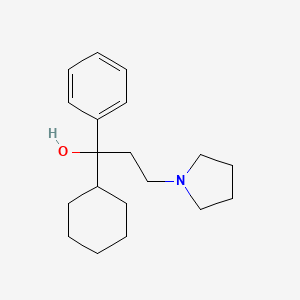 |
0.195 | ||
| ENC001230 | 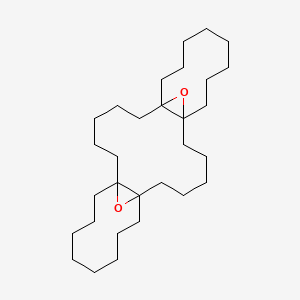 |
0.313 | D02LRQ | 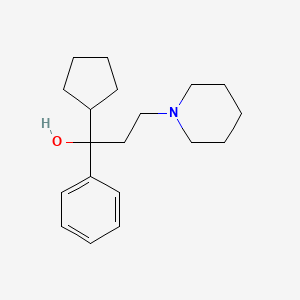 |
0.195 | ||
| ENC005710 | 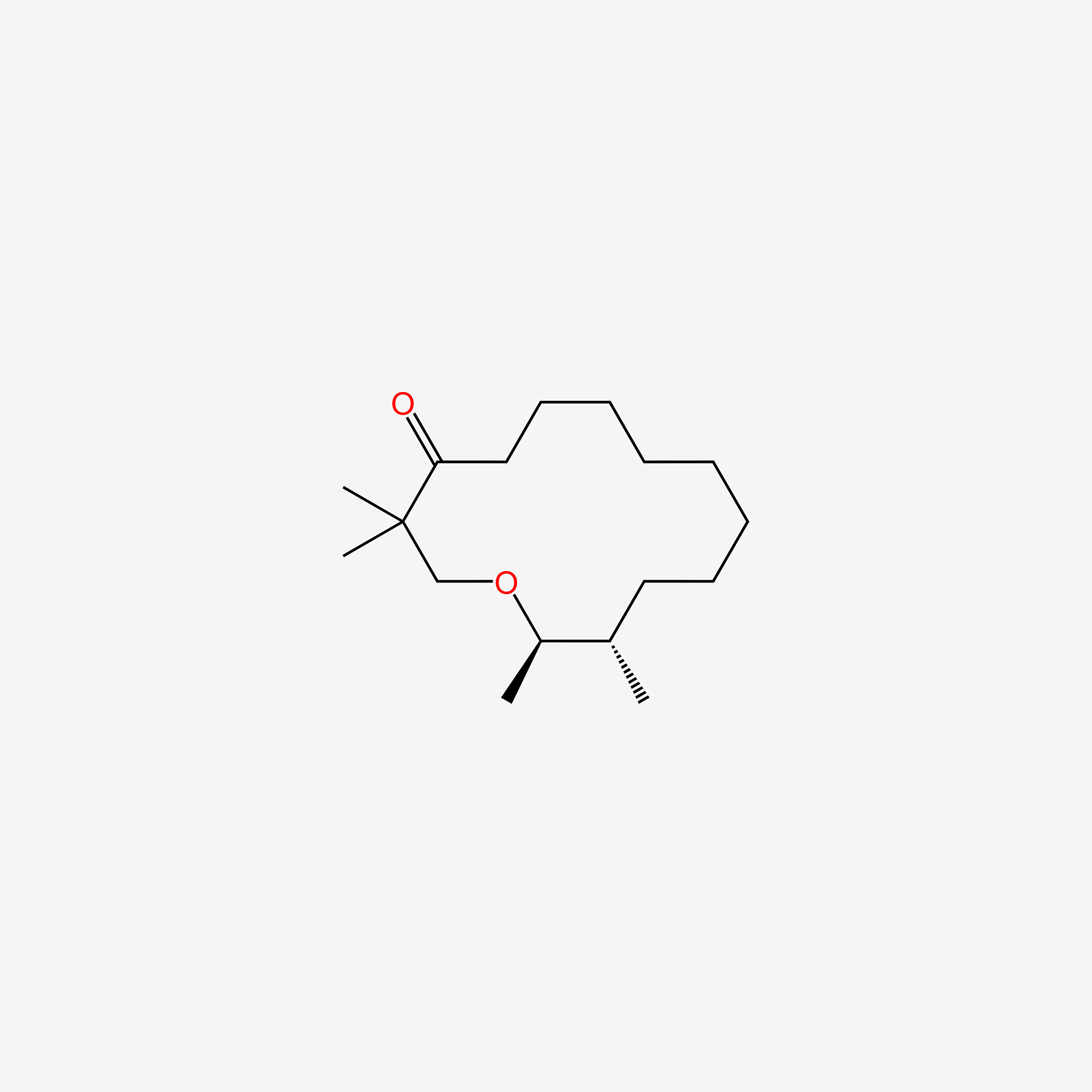 |
0.302 | D04URO | 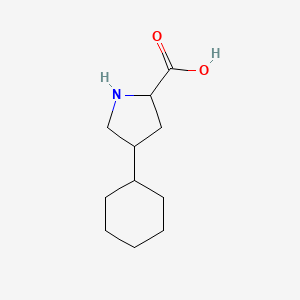 |
0.186 | ||