NPs Basic Information
|
Name |
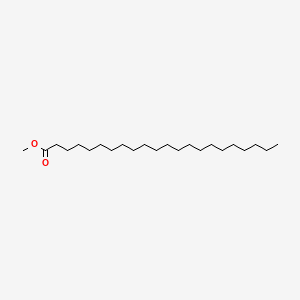
Methyl behenate
|
| Molecular Formula | C23H46O2 | |
| IUPAC Name* |
methyl docosanoate
|
|
| SMILES |
CCCCCCCCCCCCCCCCCCCCCC(=O)OC
|
|
| InChI |
InChI=1S/C23H46O2/c1-3-4-5-6-7-8-9-10-11-12-13-14-15-16-17-18-19-20-21-22-23(24)25-2/h3-22H2,1-2H3
|
|
| InChIKey |
QSQLTHHMFHEFIY-UHFFFAOYSA-N
|
|
| Synonyms |
Methyl behenate; Methyl docosanoate; 929-77-1; Docosanoic acid, methyl ester; Docosanoic acid methyl ester; Behenic acid methyl ester; BEHENIC ACID, METHYL ESTER; Benenic acid methyl ester; Docosanoic acid-methyl ester; 04KBO9R771; NSC-158426; HSDB 2724; EINECS 213-207-8; NSC 158426; BEHENICACIDMETHYLESTER; UNII-04KBO9R771; AI3-36456; MFCD00009347; Behenic Methyl Ester; Kemester 9022; docosanoic acid methyl; Docosanoic acid, methylester; SCHEMBL24669; n-Docosanoic acid methyl ester; METHYL BEHENATE [INCI]; DTXSID6029206; CDAA-251022M; CHEBI:143586; DOCOSANOIC ACID,METHYL ESTER; Methyl behenate, analytical standard; NSC158426; ZINC70455483; AKOS015851589; BEHENIC ACID METHYL ESTER [MI]; CS-W009798; HY-W009082; Docosanoic acid methyl ester (FAME MIX); BEHENIC ACID, METHYL ESTER [HSDB]; DB-057352; B1241; B1748; FT-0633117; Methyl behenate, >=98.5% (capillary GC); D88853; A854609; W-100264; Q27247655; C444918B-ABDE-4650-8507-DA7E3AAC54EB
|
|
| CAS | 929-77-1 | |
| PubChem CID | 13584 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 354.6 | ALogp: | 11.2 |
| HBD: | 0 | HBA: | 2 |
| Rotatable Bonds: | 21 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 26.3 | Aromatic Rings: | 0 |
| Heavy Atoms: | 25 | QED Weighted: | 0.157 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.047 | MDCK Permeability: | 0.00000879 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.003 | 20% Bioavailability (F20%): | 0.923 |
| 30% Bioavailability (F30%): | 1 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.06 | Plasma Protein Binding (PPB): | 97.00% |
| Volume Distribution (VD): | 3.467 | Fu: | 1.21% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.116 | CYP1A2-substrate: | 0.171 |
| CYP2C19-inhibitor: | 0.237 | CYP2C19-substrate: | 0.059 |
| CYP2C9-inhibitor: | 0.077 | CYP2C9-substrate: | 0.96 |
| CYP2D6-inhibitor: | 0.267 | CYP2D6-substrate: | 0.032 |
| CYP3A4-inhibitor: | 0.255 | CYP3A4-substrate: | 0.032 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 4.634 | Half-life (T1/2): | 0.098 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.437 | Human Hepatotoxicity (H-HT): | 0.021 |
| Drug-inuced Liver Injury (DILI): | 0.475 | AMES Toxicity: | 0.006 |
| Rat Oral Acute Toxicity: | 0.015 | Maximum Recommended Daily Dose: | 0.016 |
| Skin Sensitization: | 0.968 | Carcinogencity: | 0.036 |
| Eye Corrosion: | 0.947 | Eye Irritation: | 0.925 |
| Respiratory Toxicity: | 0.865 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000724 |  |
0.923 | D00AOJ |  |
0.738 | ||
| ENC000474 |  |
0.917 | D07ILQ |  |
0.571 | ||
| ENC000497 | 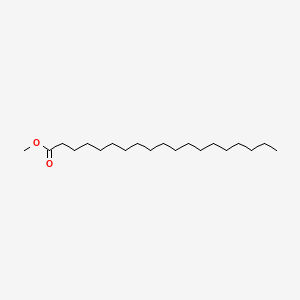 |
0.875 | D00STJ | 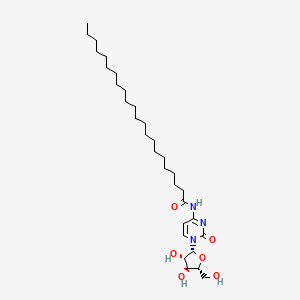 |
0.525 | ||
| ENC000553 | 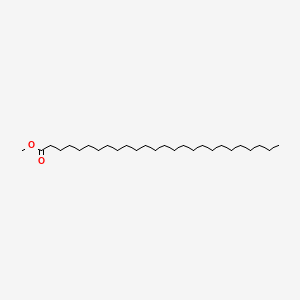 |
0.857 | D00FGR | 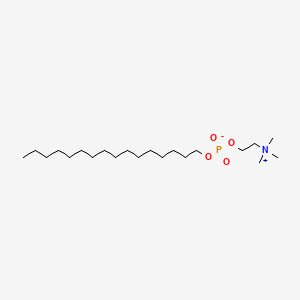 |
0.500 | ||
| ENC000280 |  |
0.833 | D0Z5SM |  |
0.465 | ||
| ENC000282 |  |
0.831 | D0O1PH |  |
0.438 | ||
| ENC000496 |  |
0.792 | D05ATI |  |
0.400 | ||
| ENC000432 |  |
0.789 | D00MLW | 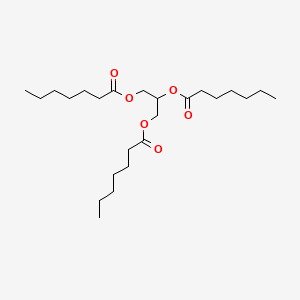 |
0.381 | ||
| ENC000765 | 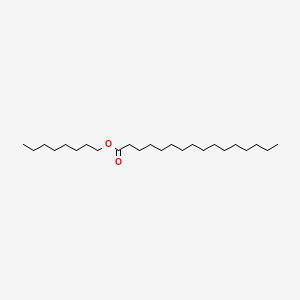 |
0.771 | D0T9TJ | 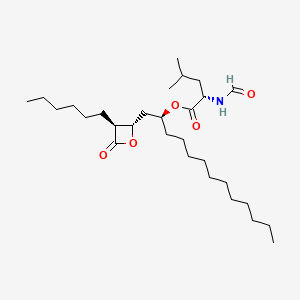 |
0.360 | ||
| ENC000761 | 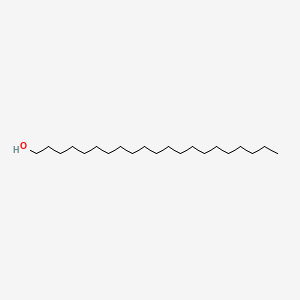 |
0.766 | D0Z1QC | 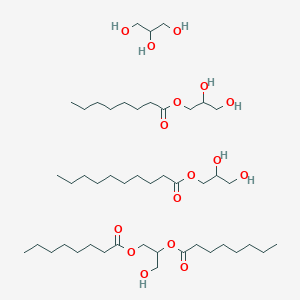 |
0.344 | ||