NPs Basic Information
|
Name |
Nonane
|
| Molecular Formula | C9H20 | |
| IUPAC Name* |
nonane
|
|
| SMILES |
CCCCCCCCC
|
|
| InChI |
InChI=1S/C9H20/c1-3-5-7-9-8-6-4-2/h3-9H2,1-2H3
|
|
| InChIKey |
BKIMMITUMNQMOS-UHFFFAOYSA-N
|
|
| Synonyms |
NONANE; n-Nonane; 111-84-2; Shellsol 140; Nonyl hydride; Heptane, ethyl-; Iotrochotin; T9W3VH6G10; CHEBI:32892; NSC-72430; nonan; DSSTox_CID_5796; Nonane, analytical standard; DSSTox_RID_77926; DSSTox_GSID_25796; 144637-82-1; MFCD02099450; 66039-00-7; CAS-111-84-2; CCRIS 6081; HSDB 107; EINECS 203-913-4; NSC 72430; UNII-T9W3VH6G10; Lodyne S; DD9; MFCD00009574; Lodyne S 100; Nonane, 99%; NONANE [HSDB]; EC 203-913-4; NONANE MFC9 H20; NONANE-5-C12; Nonane, anhydrous, >=99%; n-C9H20; CHEMBL335900; DTXSID9025796; Nonane, ReagentPlus(R), 99%; CH3-[CH2]7-CH3; NSC72430; ZINC1698517; Tox21_201479; Tox21_303148; LMFA11000579; AKOS015904046; n-Nonane 10 microg/mL in Cyclohexane; n-Nonane 1000 microg/mL in Methanol; UN 1920; NCGC00091787-01; NCGC00091787-02; NCGC00257029-01; NCGC00259030-01; LS-13716; DB-041010; DB-063623; FT-0631631; N0286; S0281; 2-ISOPROPYL-4-METHYL-6-HYDROPYRIMIDINE; Fluorochemical surfactant, anionic / non-ionic; A802420; Q150694; Fluorochemical surfactant, Zwitterionic / non-ionic; W-108667; C8F3CAB9-DAF5-4085-84EB-07C0AB04D3A1; 61193-19-9
|
|
| CAS | 111-84-2 | |
| PubChem CID | 8141 | |
| ChEMBL ID | CHEMBL335900 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 128.25 | ALogp: | 4.5 |
| HBD: | 0 | HBA: | 0 |
| Rotatable Bonds: | 6 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 0.0 | Aromatic Rings: | 0 |
| Heavy Atoms: | 9 | QED Weighted: | 0.464 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.342 | MDCK Permeability: | 0.00001440 |
| Pgp-inhibitor: | 0.002 | Pgp-substrate: | 0.002 |
| Human Intestinal Absorption (HIA): | 0.002 | 20% Bioavailability (F20%): | 0.838 |
| 30% Bioavailability (F30%): | 0.99 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.909 | Plasma Protein Binding (PPB): | 96.48% |
| Volume Distribution (VD): | 2.904 | Fu: | 3.69% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.959 | CYP1A2-substrate: | 0.545 |
| CYP2C19-inhibitor: | 0.582 | CYP2C19-substrate: | 0.57 |
| CYP2C9-inhibitor: | 0.378 | CYP2C9-substrate: | 0.899 |
| CYP2D6-inhibitor: | 0.054 | CYP2D6-substrate: | 0.121 |
| CYP3A4-inhibitor: | 0.081 | CYP3A4-substrate: | 0.106 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 5.968 | Half-life (T1/2): | 0.275 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.082 | Human Hepatotoxicity (H-HT): | 0.014 |
| Drug-inuced Liver Injury (DILI): | 0.105 | AMES Toxicity: | 0.008 |
| Rat Oral Acute Toxicity: | 0.066 | Maximum Recommended Daily Dose: | 0.025 |
| Skin Sensitization: | 0.834 | Carcinogencity: | 0.065 |
| Eye Corrosion: | 0.993 | Eye Irritation: | 0.978 |
| Respiratory Toxicity: | 0.576 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000493 |  |
0.893 | D05ATI |  |
0.385 | ||
| ENC000473 |  |
0.806 | D0Z5SM |  |
0.339 | ||
| ENC000272 |  |
0.735 | D0E4WR |  |
0.333 | ||
| ENC000421 | 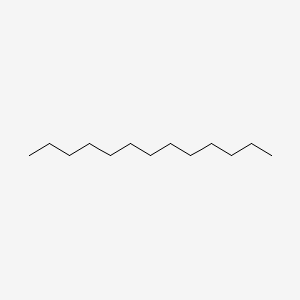 |
0.676 | D0Z5BC | 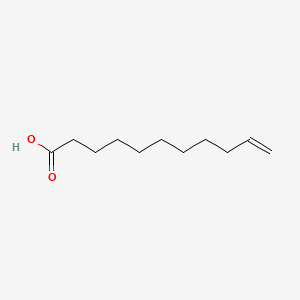 |
0.326 | ||
| ENC000049 | 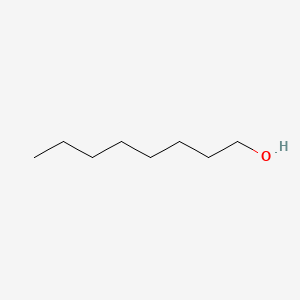 |
0.667 | D0Y8DP | 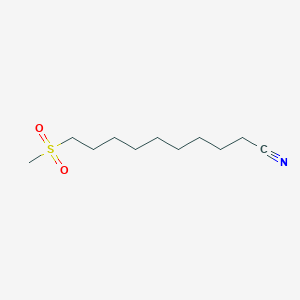 |
0.320 | ||
| ENC000279 | 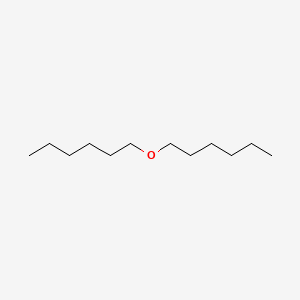 |
0.632 | D0AY9Q | 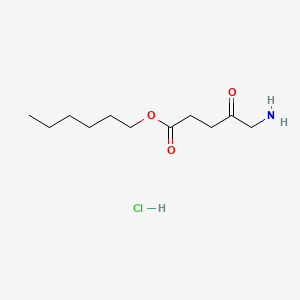 |
0.314 | ||
| ENC000422 | 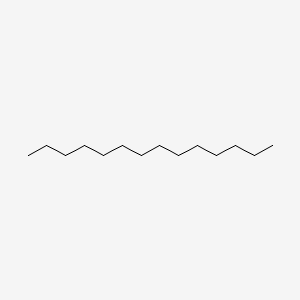 |
0.625 | D0XN8C | 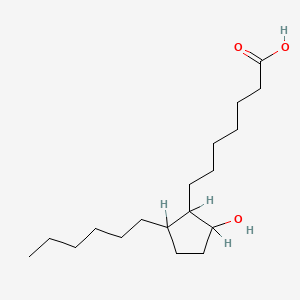 |
0.313 | ||
| ENC000451 |  |
0.618 | D03ZJE |  |
0.313 | ||
| ENC000542 |  |
0.606 | D07ILQ |  |
0.308 | ||
| ENC000317 |  |
0.606 | D01QLH | 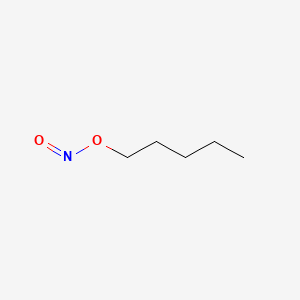 |
0.306 | ||