NPs Basic Information
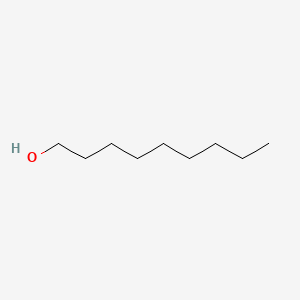
|
Name |
Nonan-1-ol
|
| Molecular Formula | C9H20O | |
| IUPAC Name* |
nonan-1-ol
|
|
| SMILES |
CCCCCCCCCO
|
|
| InChI |
InChI=1S/C9H20O/c1-2-3-4-5-6-7-8-9-10/h10H,2-9H2,1H3
|
|
| InChIKey |
ZWRUINPWMLAQRD-UHFFFAOYSA-N
|
|
| Synonyms |
1-Nonanol; Nonan-1-ol; Nonanol; 143-08-8; NONYL ALCOHOL; n-Nonyl alcohol; Pelargonic alcohol; 1-Hydroxynonane; Nonalol; Octyl carbinol; n-Nonanol; n-Nonan-1-ol; Alcohol C-9; Nonylalkohol; Pelargonalkohol; Alcohol C9; FEMA No. 2789; NSC 5521; 28473-21-4; NGK73Q6XMC; CHEBI:35986; NSC-5521; C9 alcohol; HSDB 5145; EINECS 205-583-7; UNII-NGK73Q6XMC; MFCD00002990; BRN 0969213; nonanols; nonyl-alcohol; AI3-03962; N-nonyl-alcohol; EINECS 249-048-6; Nonanol-(1); Nonyl alcohol, 8CI; 1-Nonanol, 98%; DSSTox_CID_2008; EC 205-583-7; DSSTox_RID_76457; NONYL ALCOHOL [FCC]; DSSTox_GSID_22008; SCHEMBL19807; NONYL ALCOHOL [FHFI]; NONYL ALCOHOL [HSDB]; WLN: Q9; 4-01-00-01798 (Beilstein Handbook Reference); BIDD:ER0370; CHEMBL24563; N-NONYL ALCOHOL [MI]; Pelargonic alcohol (1-nonanol); DTXSID6022008; Nonyl alcohol, >=98%, FCC; BDBM22607; FEMA 2789; Nonyl alcohol, Pelargonic alcohol; NSC5521; ZINC1686993; Tox21_300869; LMFA05000092; STL283956; AKOS009031412; CS-W009532; DB03143; 1-Nonanol, purum, >=98.0% (GC); NCGC00248194-01; NCGC00254773-01; BP-31117; BS-42231; CAS-143-08-8; FT-0608164; N0292; EN300-19921; D70513; A808013; Q161662; J-007741; F0001-0508; Z104476100; 2E051A08-F94E-40C2-88CA-7030E15C76BF
|
|
| CAS | 143-08-8 | |
| PubChem CID | 8914 | |
| ChEMBL ID | CHEMBL24563 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 144.25 | ALogp: | 4.3 |
| HBD: | 1 | HBA: | 1 |
| Rotatable Bonds: | 7 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 20.2 | Aromatic Rings: | 0 |
| Heavy Atoms: | 10 | QED Weighted: | 0.542 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.312 | MDCK Permeability: | 0.00002390 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.024 |
| Human Intestinal Absorption (HIA): | 0.003 | 20% Bioavailability (F20%): | 0.26 |
| 30% Bioavailability (F30%): | 0.967 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.974 | Plasma Protein Binding (PPB): | 86.60% |
| Volume Distribution (VD): | 1.164 | Fu: | 16.41% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.894 | CYP1A2-substrate: | 0.515 |
| CYP2C19-inhibitor: | 0.263 | CYP2C19-substrate: | 0.12 |
| CYP2C9-inhibitor: | 0.216 | CYP2C9-substrate: | 0.764 |
| CYP2D6-inhibitor: | 0.008 | CYP2D6-substrate: | 0.094 |
| CYP3A4-inhibitor: | 0.03 | CYP3A4-substrate: | 0.093 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 8.46 | Half-life (T1/2): | 0.586 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.074 | Human Hepatotoxicity (H-HT): | 0.018 |
| Drug-inuced Liver Injury (DILI): | 0.037 | AMES Toxicity: | 0.007 |
| Rat Oral Acute Toxicity: | 0.06 | Maximum Recommended Daily Dose: | 0.012 |
| Skin Sensitization: | 0.89 | Carcinogencity: | 0.089 |
| Eye Corrosion: | 0.99 | Eye Irritation: | 0.981 |
| Respiratory Toxicity: | 0.237 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000049 | 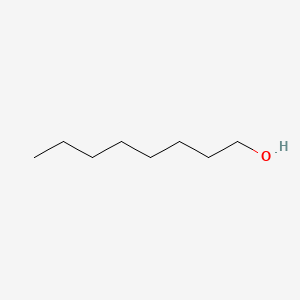 |
0.893 | D05ATI |  |
0.442 | ||
| ENC000274 | 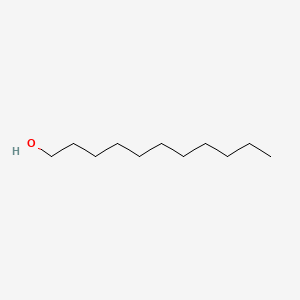 |
0.824 | D0Z5BC | 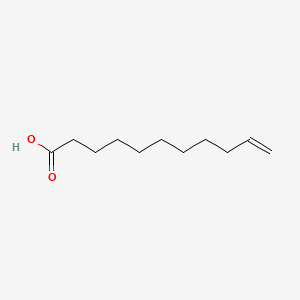 |
0.422 | ||
| ENC000276 | 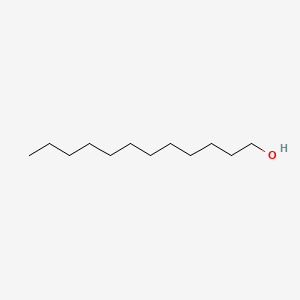 |
0.757 | D07ILQ |  |
0.419 | ||
| ENC000542 | 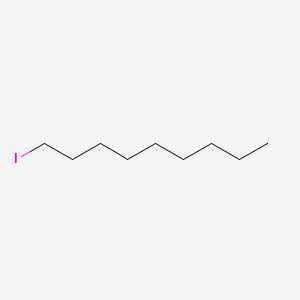 |
0.697 | D00AOJ | 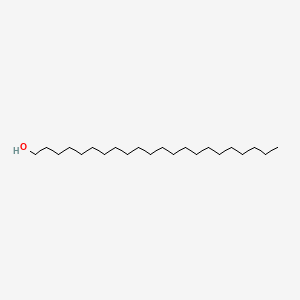 |
0.418 | ||
| ENC000493 | 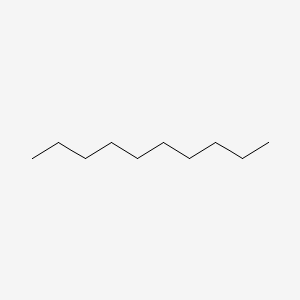 |
0.697 | D0Z5SM |  |
0.390 | ||
| ENC000330 |  |
0.697 | D0O1PH |  |
0.382 | ||
| ENC000088 |  |
0.649 | D0Y8DP |  |
0.380 | ||
| ENC000473 |  |
0.639 | D03ZJE |  |
0.359 | ||
| ENC000720 |  |
0.639 | D0XN8C |  |
0.359 | ||
| ENC000267 |  |
0.639 | D07UHS |  |
0.343 | ||