NPs Basic Information
|
Name |
Dodecane
|
| Molecular Formula | C12H26 | |
| IUPAC Name* |
dodecane
|
|
| SMILES |
CCCCCCCCCCCC
|
|
| InChI |
InChI=1S/C12H26/c1-3-5-7-9-11-12-10-8-6-4-2/h3-12H2,1-2H3
|
|
| InChIKey |
SNRUBQQJIBEYMU-UHFFFAOYSA-N
|
|
| Synonyms |
DODECANE; n-Dodecane; 112-40-3; Dihexyl; Bihexyl; Adakane 12; N-Dodecan; Undecane, methyl-; 93685-81-5; dodecan; Duodecane; Ba 51-090453; NSC 8714; 93924-07-3; CHEBI:28817; 11A386X1QH; NSC-8714; n-Dodecan [German]; D12; CCRIS 661; HSDB 5133; EINECS 203-967-9; BRN 1697175; Dodekan; normal dodecane; UNII-11A386X1QH; Normal Paraffin M; EINECS 297-629-8; EINECS 300-199-7; MFCD00008969; Norpar 13; Dodecane, 99%; DODECANE [HSDB]; DODECANE [INCI]; Dodecane-[13C12]; DSSTox_CID_6913; EC 203-967-9; EC 300-199-7; DSSTox_RID_78250; DSSTox_GSID_26913; Dodecane, analytical standard; 4-01-00-00498 (Beilstein Handbook Reference); CHEMBL30959; Density Standard 749 kg/m3; Dodecane, anhydrous, >=99%; WLN: 12H; CH3(CH2)10CH3; DTXSID0026913; NSC8714; CH3-[CH2]10-CH3; ZINC1531085; Tox21_303615; Dodecane, ReagentPlus(R), >=99%; LMFA11000004; STL280320; Dodecane, technical, >=90% (GC); AKOS015904160; NCGC00166012-01; NCGC00257481-01; CAS-112-40-3; DA-16704; LS-14163; CS-0152244; D0968; FT-0625568; C08374; Q150744; 1310FACD-F2BF-4FD7-BC20-B21DF06EDE79; J-002767; Dodecane, certified reference material, TraceCERT(R); F0001-0259; Density Standard 749 kg/m3, H&D Fitzgerald Ltd. Quality
|
|
| CAS | 112-40-3 | |
| PubChem CID | 8182 | |
| ChEMBL ID | CHEMBL30959 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 170.33 | ALogp: | 6.1 |
| HBD: | 0 | HBA: | 0 |
| Rotatable Bonds: | 9 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 0.0 | Aromatic Rings: | 0 |
| Heavy Atoms: | 12 | QED Weighted: | 0.412 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.538 | MDCK Permeability: | 0.00001170 |
| Pgp-inhibitor: | 0.005 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.002 | 20% Bioavailability (F20%): | 0.626 |
| 30% Bioavailability (F30%): | 0.996 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.429 | Plasma Protein Binding (PPB): | 97.67% |
| Volume Distribution (VD): | 3.317 | Fu: | 1.97% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.842 | CYP1A2-substrate: | 0.238 |
| CYP2C19-inhibitor: | 0.553 | CYP2C19-substrate: | 0.248 |
| CYP2C9-inhibitor: | 0.25 | CYP2C9-substrate: | 0.928 |
| CYP2D6-inhibitor: | 0.11 | CYP2D6-substrate: | 0.088 |
| CYP3A4-inhibitor: | 0.179 | CYP3A4-substrate: | 0.074 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 4.966 | Half-life (T1/2): | 0.145 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.145 | Human Hepatotoxicity (H-HT): | 0.011 |
| Drug-inuced Liver Injury (DILI): | 0.155 | AMES Toxicity: | 0.007 |
| Rat Oral Acute Toxicity: | 0.05 | Maximum Recommended Daily Dose: | 0.027 |
| Skin Sensitization: | 0.928 | Carcinogencity: | 0.049 |
| Eye Corrosion: | 0.994 | Eye Irritation: | 0.963 |
| Respiratory Toxicity: | 0.575 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000421 |  |
0.919 | D05ATI |  |
0.558 | ||
| ENC000473 |  |
0.912 | D0Z5SM |  |
0.492 | ||
| ENC000422 |  |
0.850 | D05QNO |  |
0.464 | ||
| ENC000493 |  |
0.824 | D07ILQ |  |
0.446 | ||
| ENC000423 |  |
0.791 | D0Y8DP |  |
0.415 | ||
| ENC000274 |  |
0.744 | D0O1PH |  |
0.408 | ||
| ENC000379 |  |
0.739 | D00AOJ |  |
0.403 | ||
| ENC000261 |  |
0.735 | D00FGR |  |
0.380 | ||
| ENC001127 | 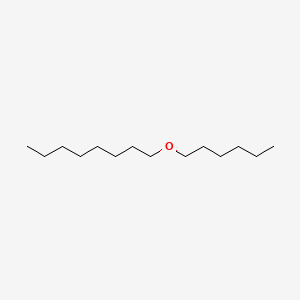 |
0.711 | D0XN8C | 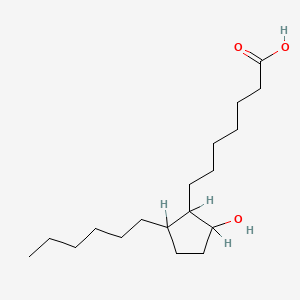 |
0.368 | ||
| ENC000399 | 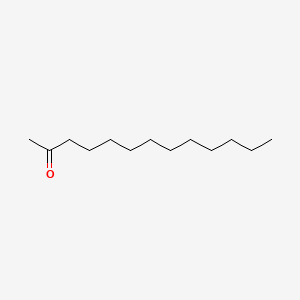 |
0.698 | D00MLW | 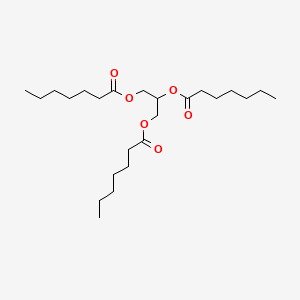 |
0.356 | ||