NPs Basic Information

|
Name |
Methyl hexanoate
|
| Molecular Formula | C7H14O2 | |
| IUPAC Name* |
methyl hexanoate
|
|
| SMILES |
CCCCCC(=O)OC
|
|
| InChI |
InChI=1S/C7H14O2/c1-3-4-5-6-7(8)9-2/h3-6H2,1-2H3
|
|
| InChIKey |
NUKZAGXMHTUAFE-UHFFFAOYSA-N
|
|
| Synonyms |
METHYL HEXANOATE; Methyl caproate; 106-70-7; Hexanoic acid, methyl ester; Hexanoic Acid Methyl Ester; Methyl hexoate; Methyl n-hexanoate; Methyl capronate; Methyl hexylate; Caproic acid methyl ester; FEMA No. 2708; Methyl n-hexoate; NSC 5023; Caproic acid, methyl ester; n-Caproic acid methyl ester; Methyl ester of hexanoic acid; HEXANOIC ACID,METHYL ESTER; 246364VPJS; NSC-5023; WE(1:0/6:0); Methyl caproate (natural); EINECS 203-425-1; BRN 1744683; methylcaproate; UNII-246364VPJS; Hexanoic acid methyl; MFCD00009510; Methyl hexanoate, 99%; hexanoic acid-methyl ester; EC 203-425-1; SCHEMBL124681; METHYL CAPROATE [INCI]; METHYL HEXANOATE [FCC]; METHYL HEXANOATE [FHFI]; DTXSID0047616; CHEBI:77322; FEMA 2708; METHYL CAPROATE [USP-RS]; Methyl hexanoate, >=99%, FG; NSC5023; ZINC1680660; LMFA07010433; Methyl hexanoate, analytical standard; STL453686; AKOS000121260; Methyl hexanoate, natural, >=99%, FG; LS-13339; FT-0628711; H0111; S0304; EN300-15456; A801486; J-001632; Q3135043; 338DD8BE-299B-44EE-816C-16C9BDD20EC6; Hexanoic acid-methyl ester 1000 microg/mL in n-Hexane; Methyl hexanoate, certified reference material, TraceCERT(R); Methyl caproate, United States Pharmacopeia (USP) Reference Standard
|
|
| CAS | 106-70-7 | |
| PubChem CID | 7824 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 130.18 | ALogp: | 2.5 |
| HBD: | 0 | HBA: | 2 |
| Rotatable Bonds: | 5 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 26.3 | Aromatic Rings: | 0 |
| Heavy Atoms: | 9 | QED Weighted: | 0.431 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.321 | MDCK Permeability: | 0.00003030 |
| Pgp-inhibitor: | 0.016 | Pgp-substrate: | 0.019 |
| Human Intestinal Absorption (HIA): | 0.003 | 20% Bioavailability (F20%): | 0.038 |
| 30% Bioavailability (F30%): | 0.723 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.997 | Plasma Protein Binding (PPB): | 53.67% |
| Volume Distribution (VD): | 0.78 | Fu: | 56.38% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.928 | CYP1A2-substrate: | 0.767 |
| CYP2C19-inhibitor: | 0.426 | CYP2C19-substrate: | 0.823 |
| CYP2C9-inhibitor: | 0.135 | CYP2C9-substrate: | 0.787 |
| CYP2D6-inhibitor: | 0.015 | CYP2D6-substrate: | 0.223 |
| CYP3A4-inhibitor: | 0.033 | CYP3A4-substrate: | 0.206 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 9.673 | Half-life (T1/2): | 0.85 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.029 | Human Hepatotoxicity (H-HT): | 0.047 |
| Drug-inuced Liver Injury (DILI): | 0.129 | AMES Toxicity: | 0.012 |
| Rat Oral Acute Toxicity: | 0.061 | Maximum Recommended Daily Dose: | 0.031 |
| Skin Sensitization: | 0.646 | Carcinogencity: | 0.253 |
| Eye Corrosion: | 0.962 | Eye Irritation: | 0.978 |
| Respiratory Toxicity: | 0.203 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000253 | 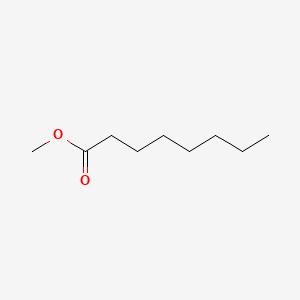 |
0.800 | D01QLH | 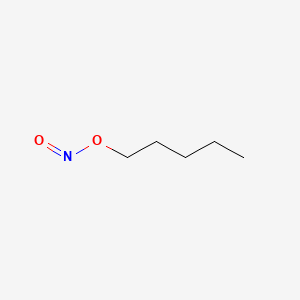 |
0.394 | ||
| ENC000249 | 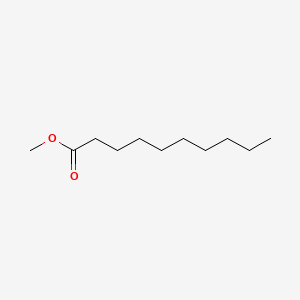 |
0.667 | D0OL6O | 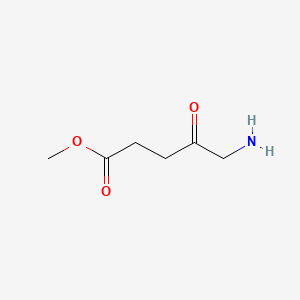 |
0.389 | ||
| ENC000250 | 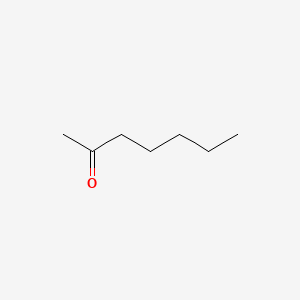 |
0.607 | D0AY9Q | 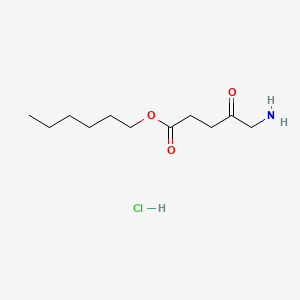 |
0.347 | ||
| ENC001036 | 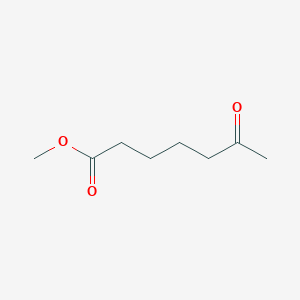 |
0.606 | D09ANG | 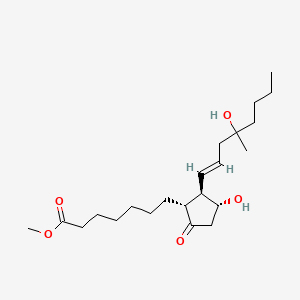 |
0.307 | ||
| ENC000260 | 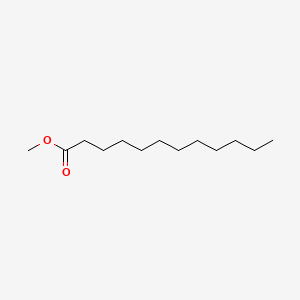 |
0.571 | D0FD0H | 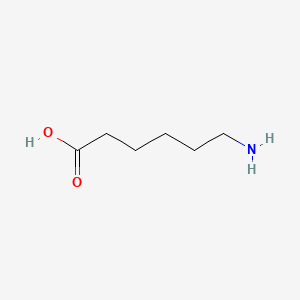 |
0.297 | ||
| ENC001253 | 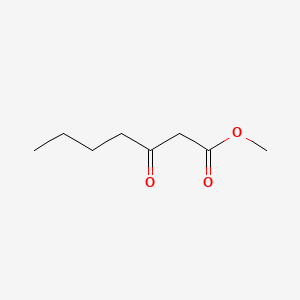 |
0.559 | D0ZI4H | 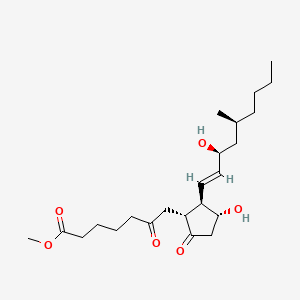 |
0.288 | ||
| ENC000228 | 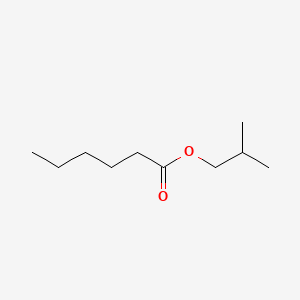 |
0.556 | D0Y3KG | 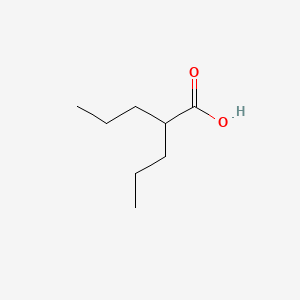 |
0.282 | ||
| ENC000315 | 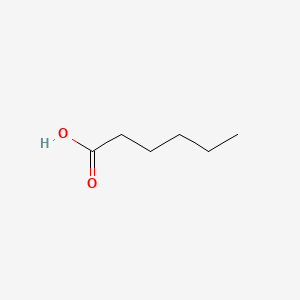 |
0.552 | D0G2KD | 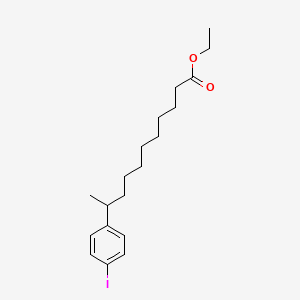 |
0.246 | ||
| ENC001025 | 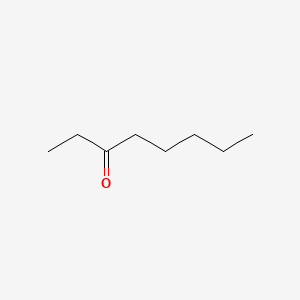 |
0.548 | D0UE9X | 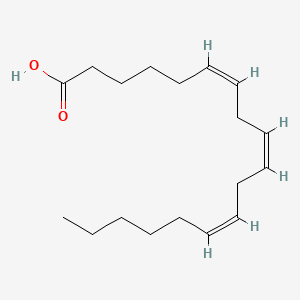 |
0.246 | ||
| ENC000254 | 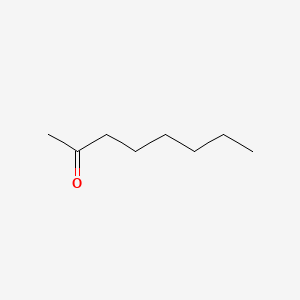 |
0.548 | D03XTC | 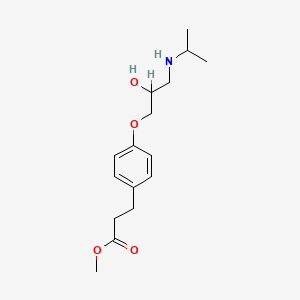 |
0.242 | ||