NPs Basic Information
|
Name |
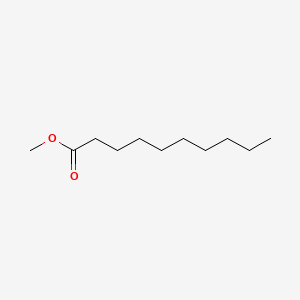
Methyl decanoate
|
| Molecular Formula | C11H22O2 | |
| IUPAC Name* |
methyl decanoate
|
|
| SMILES |
CCCCCCCCCC(=O)OC
|
|
| InChI |
InChI=1S/C11H22O2/c1-3-4-5-6-7-8-9-10-11(12)13-2/h3-10H2,1-2H3
|
|
| InChIKey |
YRHYCMZPEVDGFQ-UHFFFAOYSA-N
|
|
| Synonyms |
METHYL DECANOATE; 110-42-9; Methyl caprate; Decanoic acid, methyl ester; Methyl caprinate; Capric acid methyl ester; Methyl n-caprate; Methyl-n-caprate; Methyl n-decanoate; Uniphat A30; Metholene 2095; Decanoic acid methyl ester; decanoic acid-methyl ester; n-Capric acid methyl ester; U9L2W51J0B; DECANOIC ACID,METHYL ESTER; NSC-3713; DSSTox_CID_6842; WE(1:0/10:0); DSSTox_RID_78226; DSSTox_GSID_26842; Methyl decanoate (natural); CAS-110-42-9; CCRIS 673; HSDB 5399; NSC 3713; EINECS 203-766-6; BRN 1759170; UNII-U9L2W51J0B; AI3-26168; Decanoic acid methyl; Methyl decanoate, 99%; EC 203-766-6; CAPRIC ACID METHYLESTER; SCHEMBL122955; METHYL DECANOATE [HSDB]; CHEMBL2137647; DTXSID4026842; METHYL CAPRATE [USP-RS]; Methyl decanoate, >=99%, FG; NSC3713; CHEBI:143577; ZINC1672809; Tox21_201757; Tox21_303060; LMFA07010454; Methyl decanoate, analytical standard; MFCD00009580; AKOS009157090; CS-W016888; NCGC00163977-01; NCGC00163977-02; NCGC00256929-01; NCGC00259306-01; BS-14441; Decanoic acid, methyl ester (FAME MIX); DB-003720; D0023; FT-0628710; A802189; J-002422; Q22808395; BB30D0E8-DD7E-4A77-9781-4C29389B3F24; Methyl caprate, United States Pharmacopeia (USP) Reference Standard
|
|
| CAS | 110-42-9 | |
| PubChem CID | 8050 | |
| ChEMBL ID | CHEMBL2137647 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 186.29 | ALogp: | 4.7 |
| HBD: | 0 | HBA: | 2 |
| Rotatable Bonds: | 9 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 26.3 | Aromatic Rings: | 0 |
| Heavy Atoms: | 13 | QED Weighted: | 0.422 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.497 | MDCK Permeability: | 0.00002330 |
| Pgp-inhibitor: | 0.592 | Pgp-substrate: | 0.005 |
| Human Intestinal Absorption (HIA): | 0.003 | 20% Bioavailability (F20%): | 0.975 |
| 30% Bioavailability (F30%): | 0.971 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.985 | Plasma Protein Binding (PPB): | 91.74% |
| Volume Distribution (VD): | 0.587 | Fu: | 8.54% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.972 | CYP1A2-substrate: | 0.684 |
| CYP2C19-inhibitor: | 0.709 | CYP2C19-substrate: | 0.602 |
| CYP2C9-inhibitor: | 0.516 | CYP2C9-substrate: | 0.898 |
| CYP2D6-inhibitor: | 0.026 | CYP2D6-substrate: | 0.126 |
| CYP3A4-inhibitor: | 0.182 | CYP3A4-substrate: | 0.134 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 7.809 | Half-life (T1/2): | 0.71 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.076 | Human Hepatotoxicity (H-HT): | 0.031 |
| Drug-inuced Liver Injury (DILI): | 0.185 | AMES Toxicity: | 0.007 |
| Rat Oral Acute Toxicity: | 0.043 | Maximum Recommended Daily Dose: | 0.024 |
| Skin Sensitization: | 0.926 | Carcinogencity: | 0.204 |
| Eye Corrosion: | 0.959 | Eye Irritation: | 0.978 |
| Respiratory Toxicity: | 0.698 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000260 |  |
0.857 | D0Z5BC |  |
0.469 | ||
| ENC000253 |  |
0.833 | D05ATI |  |
0.431 | ||
| ENC000495 | 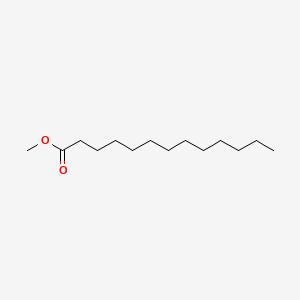 |
0.800 | D0G2KD | 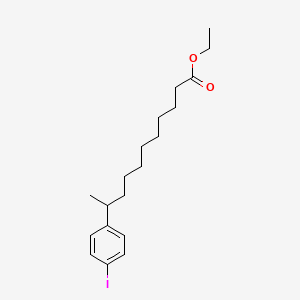 |
0.420 | ||
| ENC000604 | 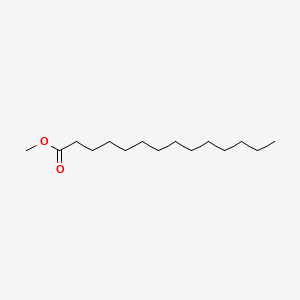 |
0.750 | D0AY9Q | 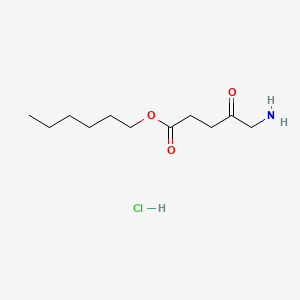 |
0.418 | ||
| ENC000248 | 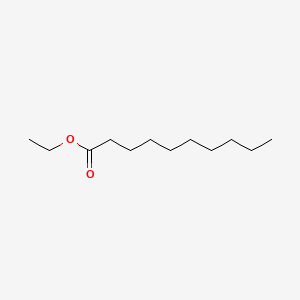 |
0.744 | D03ZJE | 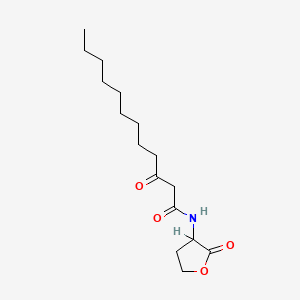 |
0.418 | ||
| ENC000265 | 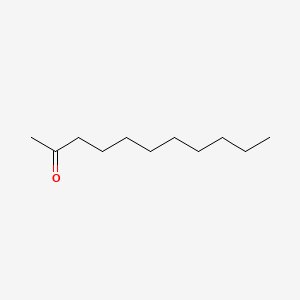 |
0.725 | D07ILQ |  |
0.412 | ||
| ENC001274 | 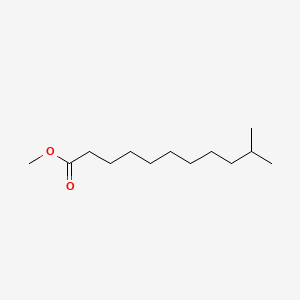 |
0.711 | D09ANG | 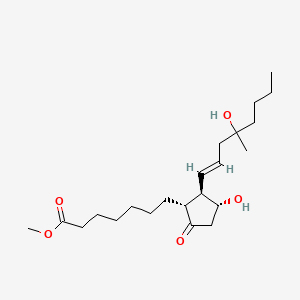 |
0.410 | ||
| ENC000560 | 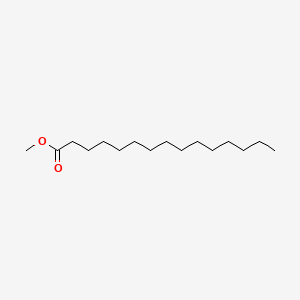 |
0.706 | D0XN8C | 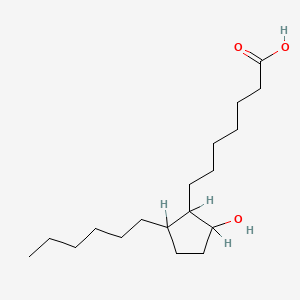 |
0.397 | ||
| ENC000259 | 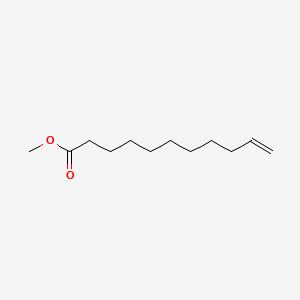 |
0.705 | D0E4WR | 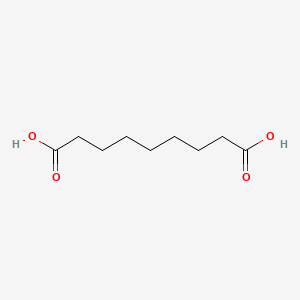 |
0.392 | ||
| ENC000722 | 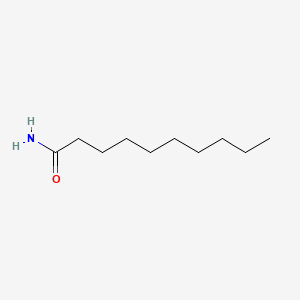 |
0.683 | D0Z5SM |  |
0.385 | ||