NPs Basic Information
|
Name |
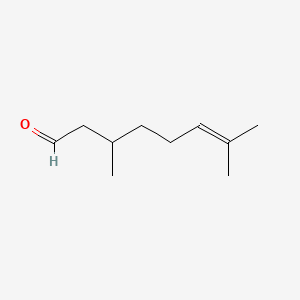
Citronellal
|
| Molecular Formula | C10H18O | |
| IUPAC Name* |
3,7-dimethyloct-6-enal
|
|
| SMILES |
CC(CCC=C(C)C)CC=O
|
|
| InChI |
InChI=1S/C10H18O/c1-9(2)5-4-6-10(3)7-8-11/h5,8,10H,4,6-7H2,1-3H3
|
|
| InChIKey |
NEHNMFOYXAPHSD-UHFFFAOYSA-N
|
|
| Synonyms |
CITRONELLAL; 106-23-0; 3,7-Dimethyloct-6-enal; 3,7-Dimethyl-6-octenal; Rhodinal; 6-Octenal, 3,7-dimethyl-; 2,3-Dihydrocitral; (+/-)-Citronellal; Citronellel; beta-Citronellal; 3,7-Dimethyl-6-octen-1-al; 8000-29-1; CITRONELLOL,(D); .beta.-Citronellal; FEMA No. 2307; NSC 46106; CHEBI:47856; QB99VZZ7GZ; 3,7-dimethyl-oct-6-enal; 3,7-dimethyloct-6-en-1-al; NSC46106; NSC-46106; Citronella; D-Rhodinal; 6-Octenal, 3,7-dimethyl-, (R)-; Rhodinal (VAN); Citronella (natural); HSDB 594; EINECS 203-376-6; UNII-QB99VZZ7GZ; AI3-00203; CCRIS 8421; beta -Citronellal; racemic citronellal; (RS)-citronellal; (+/-)-3,7-Dimethyl-6-octenal; MFCD00038090; (+/-) citronellal; ( inverted exclamation markA)-Citronellal; 6-Octenal,7-dimethyl-; CITRONELLAL [MI]; CITRONELLAL [FCC]; CITRONELLAL [FHFI]; CITRONELLAL [HSDB]; CITRONELLAL [INCI]; Epitope ID:112868; EC 203-376-6; DSSTox_CID_21790; DSSTox_RID_79843; DSSTox_GSID_41790; SCHEMBL29275; CHEMBL447944; GTPL6300; DTXSID3041790; FEMA 2307; HY-N7126; Tox21_301195; c1026; s5585; AKOS000121405; (+/-)-Citronellal, >=85%, FG; CCG-266248; CS-W010931; (+/-)-Citronellal, analytical standard; NCGC00248328-01; NCGC00255093-01; AS-54400; CAS-106-23-0; (+/-)-Citronellal, >=95.0% (GC); DB-040680; DB-072650; C3648; FT-0604386; FT-0623961; FT-0623963; FT-0699573; EN300-21458; C17384; P50009; (+/-)-Citronellal, natural, >=85%, FCC, FG; A801397; J-502100; W-108770; Q61651085; Z104497952
|
|
| CAS | 106-23-0 | |
| PubChem CID | 7794 | |
| ChEMBL ID | CHEMBL447944 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 154.25 | ALogp: | 3.0 |
| HBD: | 0 | HBA: | 1 |
| Rotatable Bonds: | 5 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 17.1 | Aromatic Rings: | 0 |
| Heavy Atoms: | 11 | QED Weighted: | 0.435 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.407 | MDCK Permeability: | 0.00002340 |
| Pgp-inhibitor: | 0.023 | Pgp-substrate: | 0.002 |
| Human Intestinal Absorption (HIA): | 0.006 | 20% Bioavailability (F20%): | 0.815 |
| 30% Bioavailability (F30%): | 0.108 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.99 | Plasma Protein Binding (PPB): | 68.87% |
| Volume Distribution (VD): | 3.407 | Fu: | 18.17% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.578 | CYP1A2-substrate: | 0.406 |
| CYP2C19-inhibitor: | 0.116 | CYP2C19-substrate: | 0.669 |
| CYP2C9-inhibitor: | 0.037 | CYP2C9-substrate: | 0.728 |
| CYP2D6-inhibitor: | 0.049 | CYP2D6-substrate: | 0.269 |
| CYP3A4-inhibitor: | 0.033 | CYP3A4-substrate: | 0.188 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 13.045 | Half-life (T1/2): | 0.418 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.012 | Human Hepatotoxicity (H-HT): | 0.508 |
| Drug-inuced Liver Injury (DILI): | 0.03 | AMES Toxicity: | 0.024 |
| Rat Oral Acute Toxicity: | 0.011 | Maximum Recommended Daily Dose: | 0.021 |
| Skin Sensitization: | 0.961 | Carcinogencity: | 0.491 |
| Eye Corrosion: | 0.976 | Eye Irritation: | 0.987 |
| Respiratory Toxicity: | 0.859 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000229 | 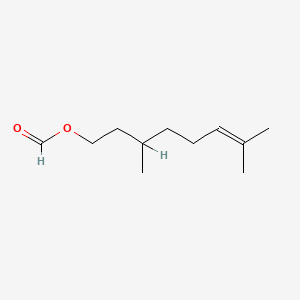 |
0.600 | D0M1PQ | 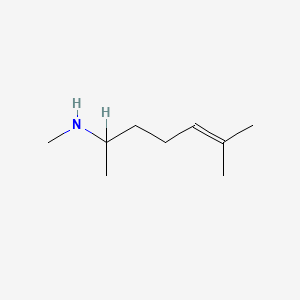 |
0.486 | ||
| ENC000311 | 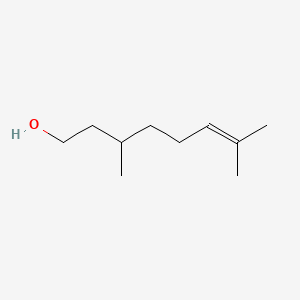 |
0.568 | D0ZK8H | 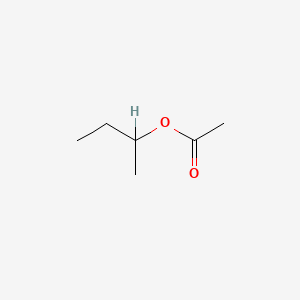 |
0.225 | ||
| ENC000319 | 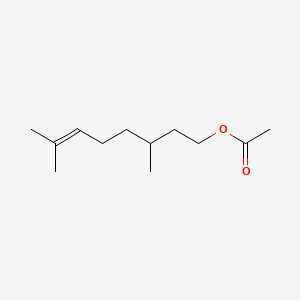 |
0.500 | D09XWD | 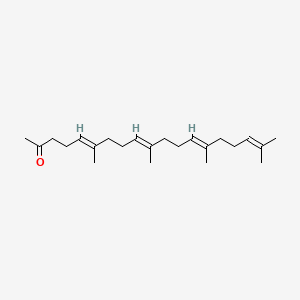 |
0.221 | ||
| ENC003366 | 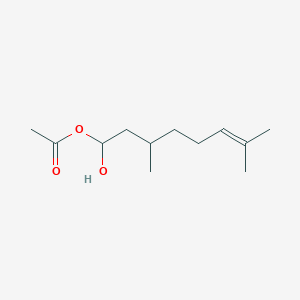 |
0.478 | D05XQE | 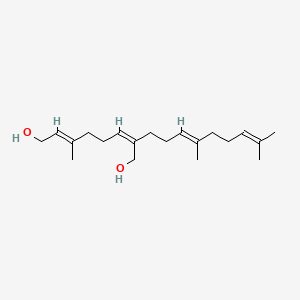 |
0.219 | ||
| ENC001150 | 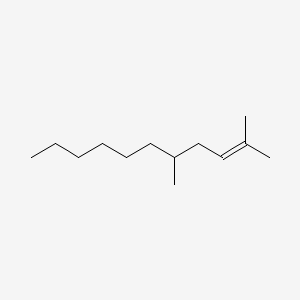 |
0.422 | D0Y3KG | 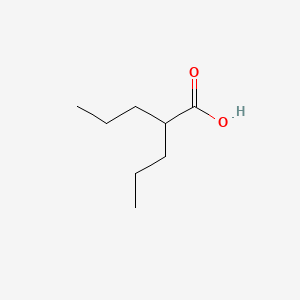 |
0.196 | ||
| ENC001434 | 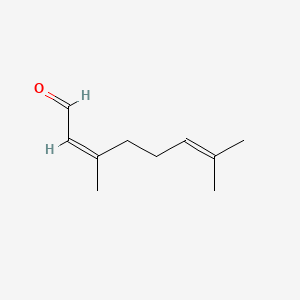 |
0.415 | D04MWJ | 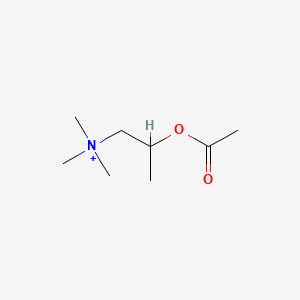 |
0.191 | ||
| ENC001424 | 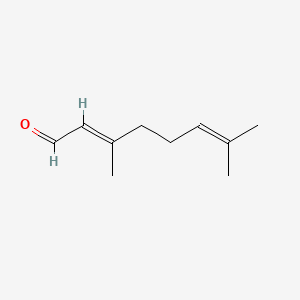 |
0.415 | D00WUF | 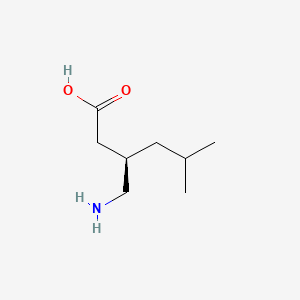 |
0.188 | ||
| ENC001649 | 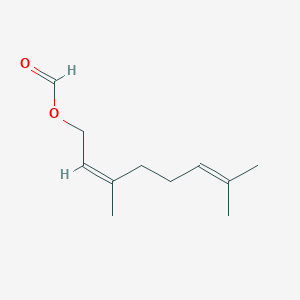 |
0.391 | D03VFL | 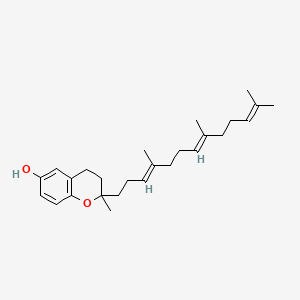 |
0.176 | ||
| ENC001718 | 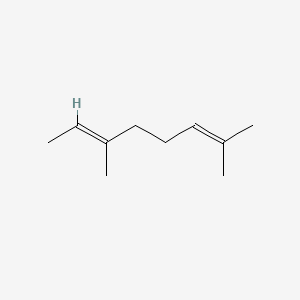 |
0.375 | D0Q9HF | 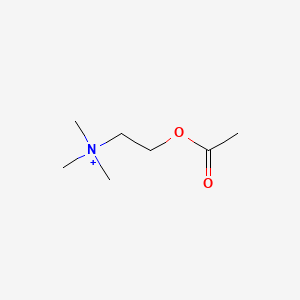 |
0.174 | ||
| ENC000846 | 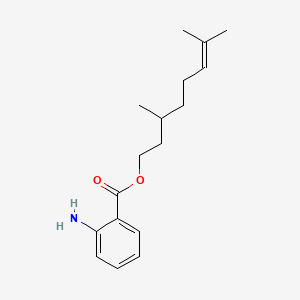 |
0.355 | D0Q6DX | 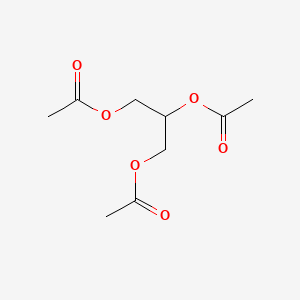 |
0.172 | ||